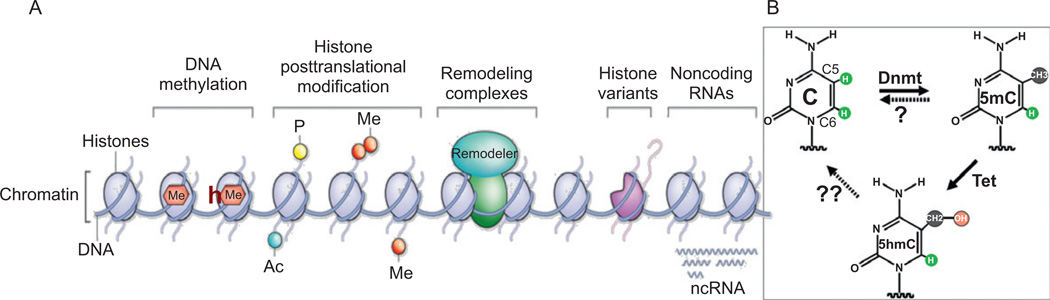
Fig. 1.
Possible components of an epigenetic “code.” (A) Five broad and interrelated phenomena affect chromatin structure: DNA methylation, histone modification, chromatin-remodeling, histone replacement by variants, and the effects of noncoding RNAs. All five have been shown to be essential contributors to the epigenetics, though DNA methylation and histone modifications have, so far, been much more extensively investigated. Adopted and modified from Ref. 3. (B) DNA cytosine methylation, hydroxylation, and demethylation. The question mark indicates possible activity of DNA demethylases.4–9 “Tet” indicates conversion of 5mC to 5hmC in mammalian DNA, by the MLL fusion partner TET1.10 It is currently unknown whether 5hmC is an end product or an intermediate in active DNA demethylation. The double question mark indicates a possible MTase-assisted removal of the C5-bound hydroxymethyl group.11