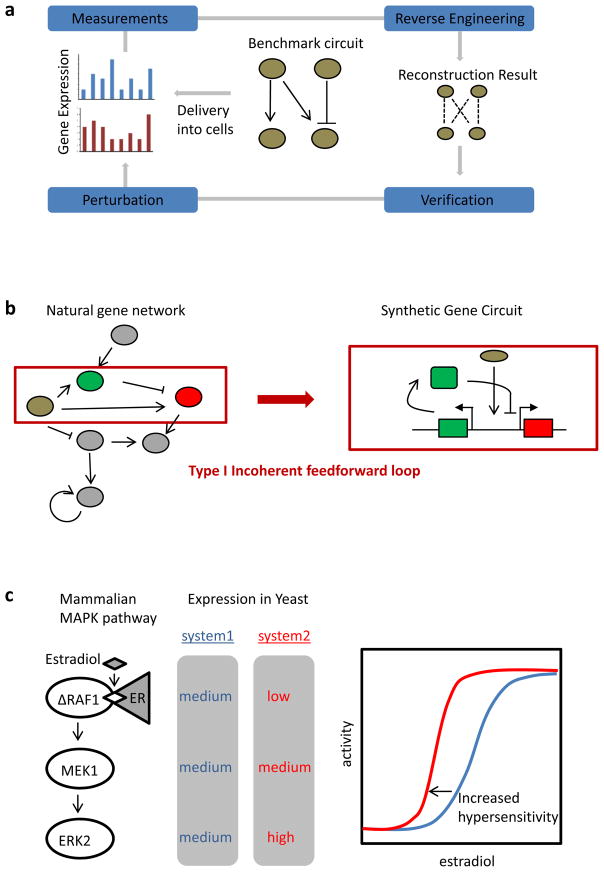
Figure 3. Studying gene and signalling networks.
a | Schematic of reverse engineering with synthetic circuits. A benchmark gene circuit is integrated into a mammalian cell line. Different reverse engineering algorithms are used to generate a network model from perturbation and gene expression analysis. The quality of reverse engineering algorithms is assessed and improved by comparing the resulting models to the actual circuit architecture in an iterative cycle93. b | Studying gene network architectures. Synthetic gene circuits enable the study of network modules in isolation from their natural context in which they may be interconnected with other endogenous signalling pathways (shown in blue). The schematic shows the design of a type I incoherent feedforward loop circuit, consisting of an input (shown in brown) that activates both an output (shown in red) and an 97 heterologous reconstitution. A mammalian MAPK pathway, consisting of an estradiol-inducible RAF1 protein that activates MEK1, which in turn activates ERK2, has been expressed in yeast. Increased concentration of each protein at each step of the cascade has been shown to augment the degree of input ultrasensitivity101. ER, oestrogen receptor.