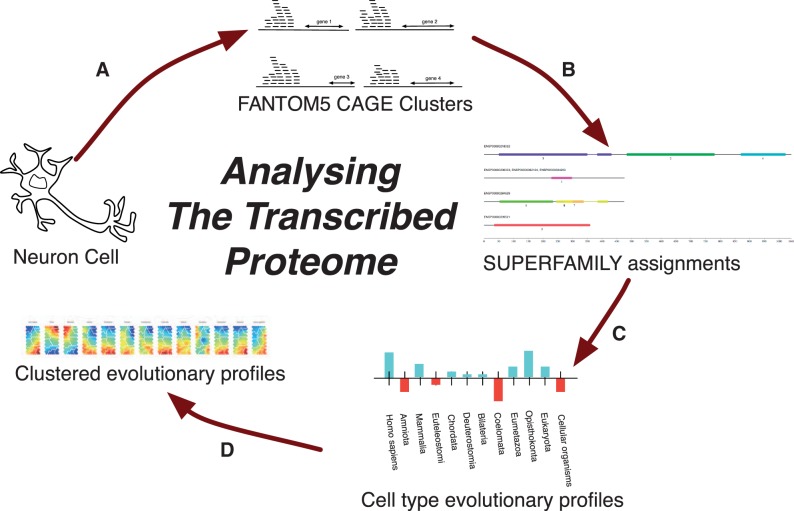
Fig. 5.
The pipeline used in presented work: (A) Clustered CAGE tags are aligned to the human genome and mapped to the closest known gene (where possible). (B) When this gene is protein coding, SUPERFAMILY is used to annotate it with its protein domain architecture. (C) Each expressed domain architecture then has its MRCA calculated, which allows for a profile of structural innovation usage for each cell type to be built up (fig. 3). (D) The profiles can then be clustered to find cell types which share a common evolutionary past. These clusters can then be analyzed to discover domain architectures key to bifurcations in cell phenotype.