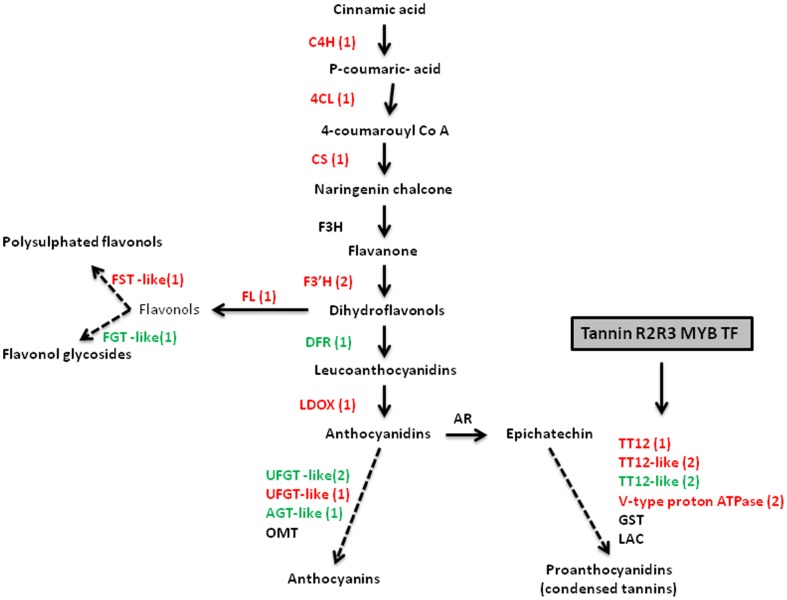
Figure 3. Predicted flavonoid pathway in cork oak ECM roots.
Enzymes are indicated by capital letters; red color indicates transcripts that were up-regulated in ECM roots compared with non-symbiotic roots. Green indicates down-regulated transcripts. Numbers in brackets indicate the number of differentially expressed genes found in the present study. Dashed arrows indicate steps which are not yet fully understood. Grey box indicates transcriptional regulators. Abbreviations are as follows: AGT anthocyanidin -o-glucosyltransferase, ANR anthocyanidin reductase, CHS chalcone synthase, C4H cinnamate 4- hydroxylase, 4CL 4-coumarate-CoA ligase, DFR dihydroflavonol 4-reductase, F3 H flavanone 3b-hydroxylase, F3′H flavonoid 3′-hydroxylase, FST flavonol sulphotransferase, FGT flavonol glucosyltransferase, GST glutathione S transferase, LAC laccase-like, LDOX leucoanthocyanidin dioxygenase, OMT O-methyltransferase, tannin R2R3 MYB TF tannin-related R2R3 MYB transcription factor, V-type H+ ATPase encoding a vacuolar proton pump that supports MATE antiporter, TT12 transparent testa 12 enconding a multidrug and toxic compound extrusion-type (MATE) transporter, UFGT UDP-glucose:flavonoid-3-O-glycosyltransferase.