Figure 4.
RECQL5 Loss Increases Transcript Elongation Rates across the Genome
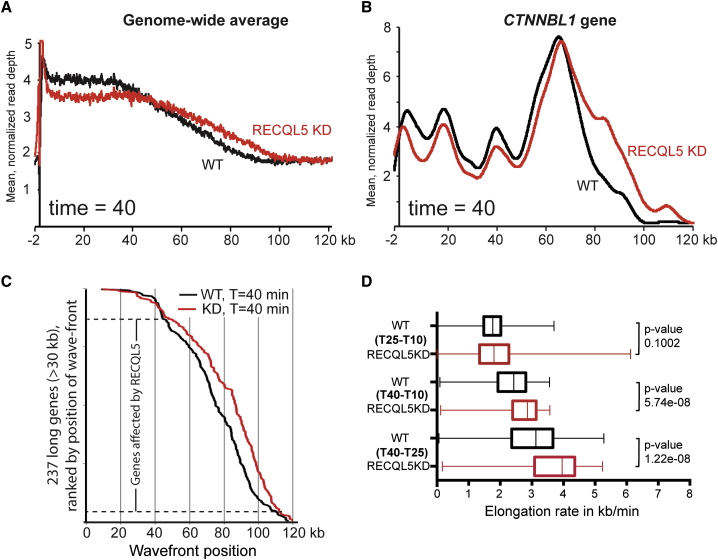
(A) Profile of normalized GRO-seq reads across the human genome 40 min after release from DRB-inhibition.
(B) Normalized and smoothed spline profile across the CTNNBL1 gene. The read-depth differs across the gene, giving rise to apparently uneven activity distribution.
(C) Positions of RNAPII wave fronts calculated over appropriately long genes 40 min after DRB removal. p value for differences between data sets <10−4.
(D) Transcription elongation rates in the indicated time intervals calculated from the position of RNAPII activity wave fronts. All genes included had to be long enough for transcript elongation to not have reached the TTS at the time of measurement, so fewer genes could be tested at later time points (see Extended Experimental Procedures). Box-plot representation shows median values ± 25% quartiles in the box and minimum/maximum distribution of the values in the whiskers. See also Figure S2.