Figure S1.
RECQL5 Levels Affect RNAPII Profiles Genome-wide, but Have Little Effect on Gene Expression Levels, Related to Figures 1 and 2
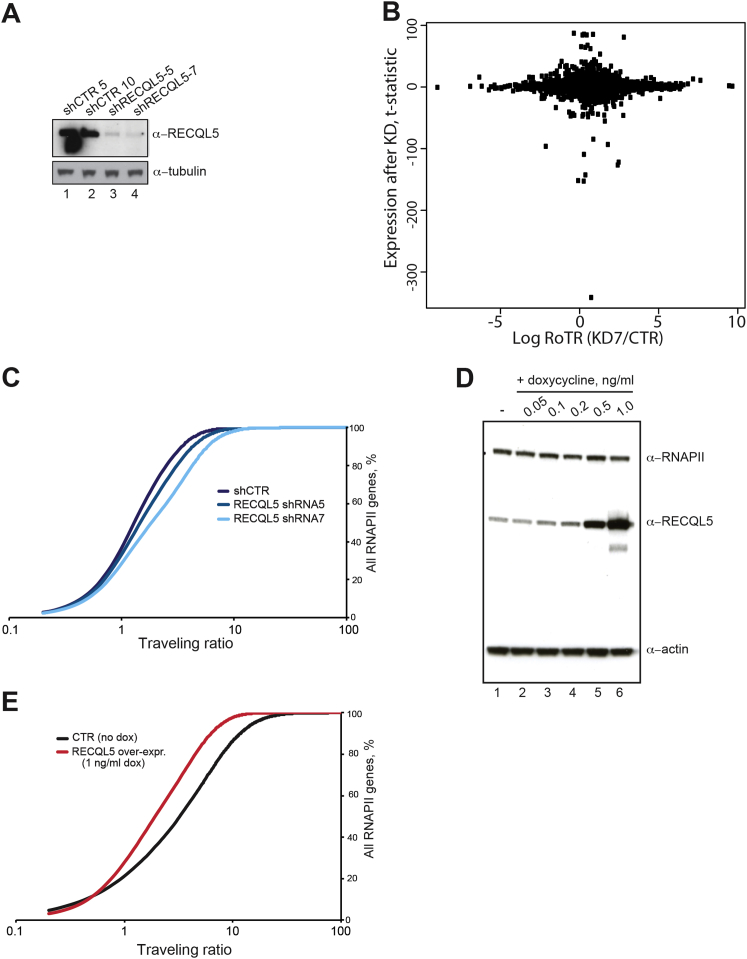
(A) Western Blot analysis showing the efficiency of the two shRNAs against RECQL5, with tubulin as loading control. Both experimental shRNAs knock down RECQL5 efficiently, but shRNA7 consistently repeatedly worked better than shRNA5 (lanes 3 and 4). Note that shCTR5 and shCTR10 are two doses of viral supernatant of the control (CTR) shRNA, showing that the infection procedure itself does not affect RECQL5 protein levels (lanes 1 and 2).
(B) Correlation plot, between gene-specific changes in the RNAPII profile (x axis; ratio of traveling ratios, RoTR), and changes in the expression of these genes (y axis; t-statistic of expression changes) upon RECQL5 knockdown. As suggested by this plot, there is little change in expression values between wild-type and knockdown (y axis). See also Table S1. More importantly, there is no correlation between genes whose expression change and those at which there is a change in promoter-proximal peak density (relative to density in coding sequence; traveling ratio) (p value > 0.7; and Spearman’s correlation −0.05).
(C) RECQL5 knockdown induces a shift in the traveling ratio. Traveling ratios were calculated for all the Ensembl transcripts and plotted, sorted according to their values. The plots show that upon RECQL5 knockdown the traveling ratios generally shift toward higher values, supporting the finding that the increased RNAPII density on the TSS and the decreased density of RNAPII in the body of the gene is a general feature.
(D) Western Blot showing RECQL5 levels with increasing doses of doxycycline. Cells were treated for 24h at the indicated concentrations of doxycycline before being harvested. RNAPII and actin are used as loading controls.
(E) As in (C), but after overexpression of RECQL5, showing that the traveling ratios generally shift toward lower values, supporting the finding that the decreased RNAPII density on the TSS and the increased density of RNAPII in the body of the gene is a general feature.