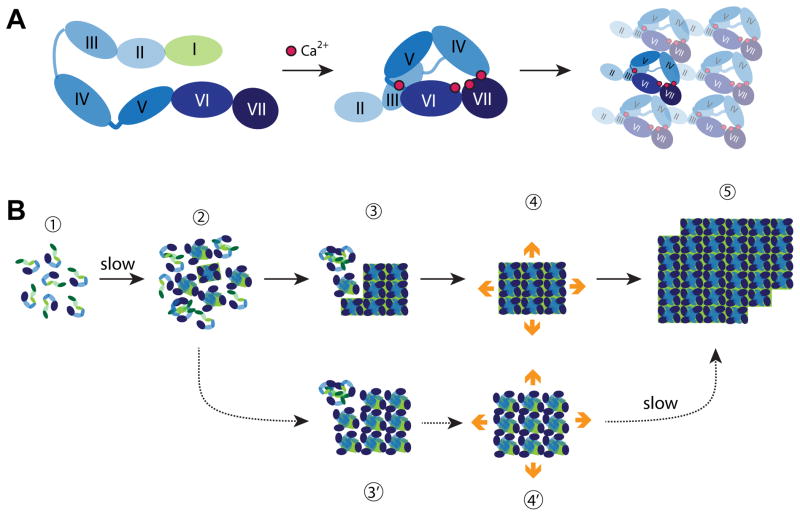
Fig. 2.
Building S-layers. (A) Schematic structure and assembly pathway of the G. stearothermophilus SbsB S-layer protein. The cell-wall attachment domain (light green) and six consecutive immunoglobulin-like domains (shades of blue) are schematically depicted in the extended monomer structure (left). The four calcium ions stabilizing intra- and inter-domain contacts are shown as red spheres in the assembly-competent monomer (middle). The cell-wall attachment domain is not resolved in the crystal structure and thus not shown. Structure of the oblique array formed upon assembly of compact monomers viewed from the outside of the cell (right). All drawings are based on reference [19••]. (B) Growth of L. sphaericus SbpA at interfaces. Monomers are believed to consist of three cell wall anchoring domains (green) followed by a calcium binding domain and three C-terminal immunoglobulin-like domains (shades of blue) [24]. Steps involved in S-layer crystallization on supported lipid bilayers are indicated with solid arrows and labeled 1 to 5. S-layers are viewed from their external side. Mica can stabilize a subpopulation of less compact but yet crystalline clusters (3′ and 4′) that eventually convert to the compact and lower energy form (step 5). All drawings based on references [22••] and [23].