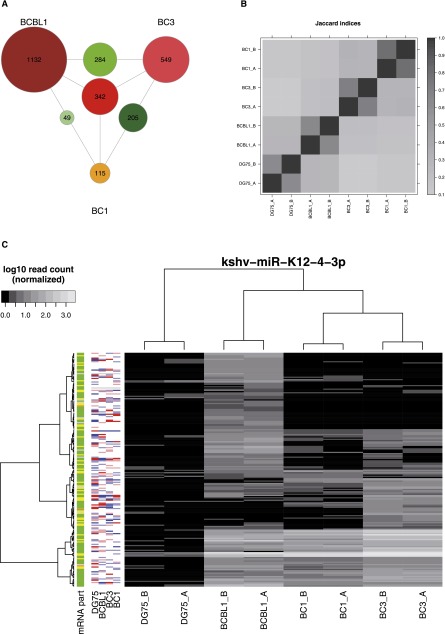
Figure 2.
Comparison of PAR-CLIP data sets. (A) The number of target sites observed only in individual cell lines (outermost labeled circles), in two cell lines (circles on the edges between cell lines), and in all three cell lines (center circle), for KSHV microRNA target sites. Relatively few target sites appear to be active in multiple cell lines (see Supplemental Fig. S2A for overlaps of cellular microRNA targets). (B) Summary of all pairwise overlaps for clusters of cellular and viral microRNAs in all data sets. The Jaccard index (J) is the number of clusters in the intersection divided by the total number of clusters in any of the two experiments. Jaccard indices of ∼70% for all replicate measurements indicate high reproducibility, whereas comparisons across cell lines show relatively low overlap (J < 40%) (see also Supplemental Fig. S2). (C) The PAR-CLIP read heatmap for target sites of the KSHV microRNA miR-K12-4-3p (see Fig. 1B for more information about PAR-CLIP read heatmaps). Between KSHV-positive cell lines, there is no correlation, but there are distinct clusters of target sites. No obvious dependency between clusters and mRNA expression level is observable.