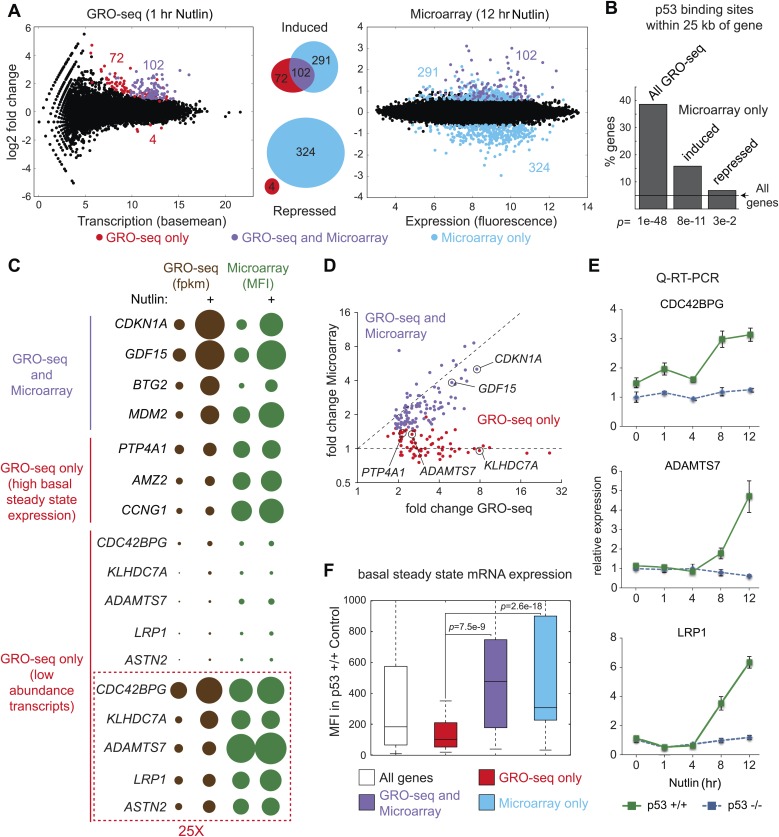
Figure 2. Global analysis of p53 effects on RNA synthesis vs steady state levels.
(A) MAplots for GRO-seq and microarray gene profiling experiments in HCT116 p53 +/+ cells after 1 hr and 12 hr of Nutlin treatment, respectively. Colors indicate whether genes scored as statistically different in both platforms (purple), in the GRO-seq only (red) or the microarray experiment only (blue). (B) Few genes downregulated in the microarray experiment show p53 binding within 25 kb of the gene, suggestive of indirect regulation. (C) Bubble plots displaying relative signals derived from the GRO-seq and microarray experiments illustrate how genes with very high basal expression or very low transcription are not significantly affected at the steady state level as measured by microarray. For the CDC42BPG, KLHDC7A, ADAMTS7, LRP1 and ASTN2 loci, the signals were replotted at 25-fold magnification. (D) Scatter plot showing comparative fold induction for p53 target genes transactivated at 1 hr Nutlin treatment between the GRO-seq and microarray experiments. (E) Q-RT-PCR indicates that many low abundance transcripts upregulated by GRO-seq are indeed induced at the steady state level. (F) Box and whisker plots showing the expression of various gene sets as detected by microarray.