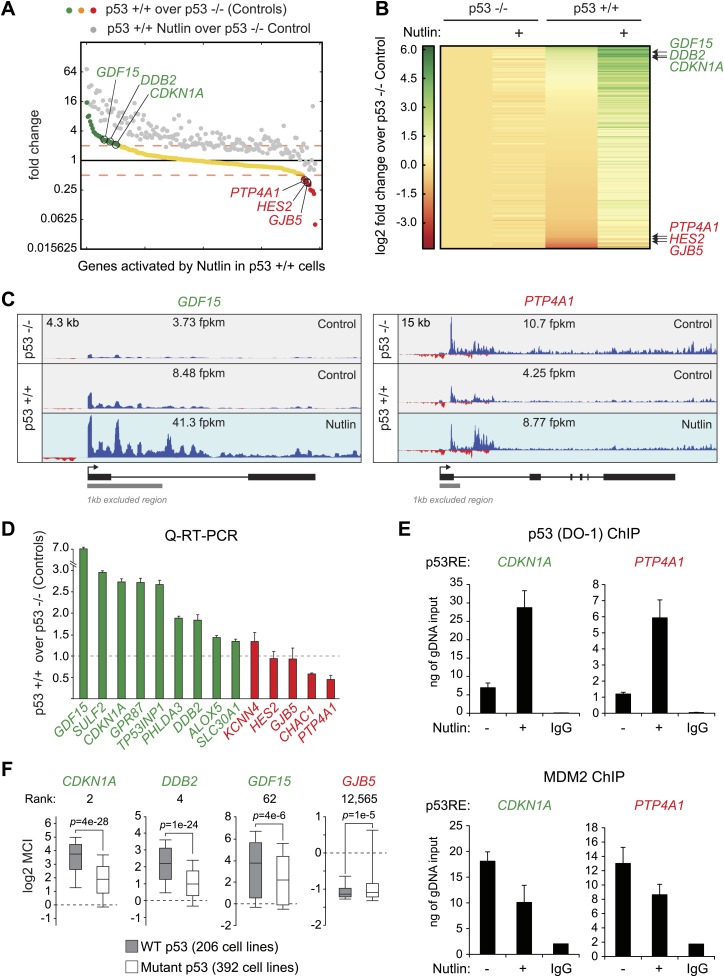
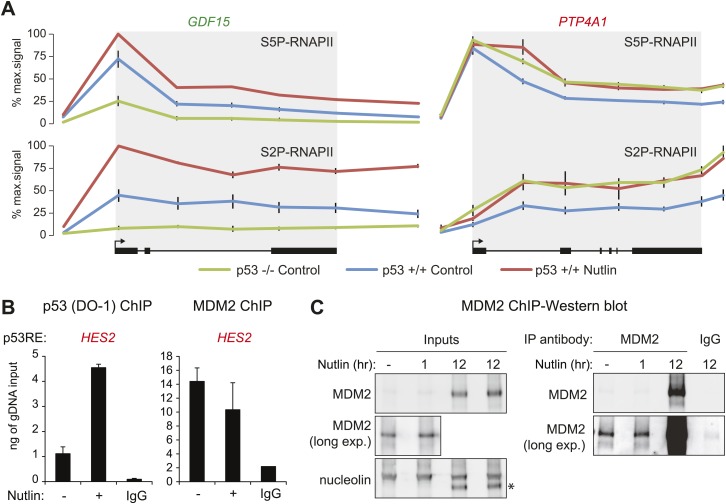
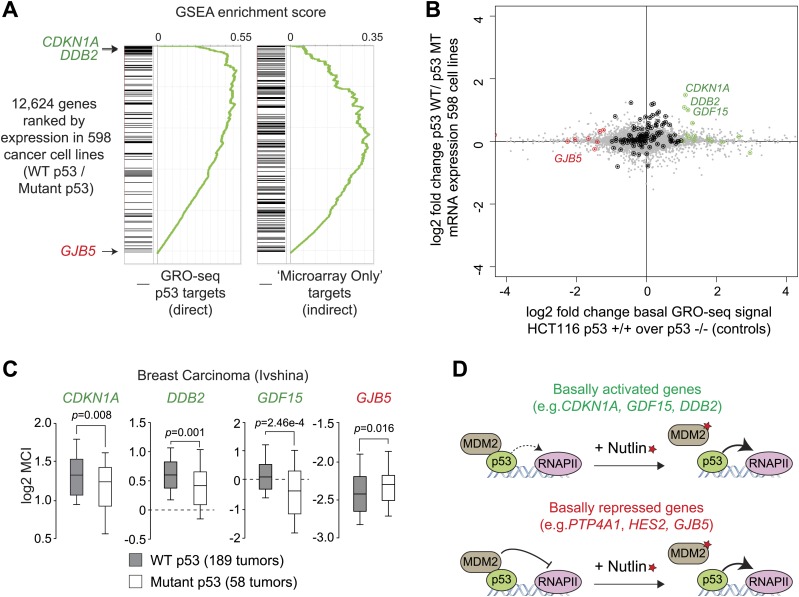
Figure 3. p53 exerts varying activating and repressing effects on its target genes prior to MDM2 inhibition.
(A) 198 genes activated upon 1 hr Nutlin treatment in HCT116 p53 +/+ cells are ranked from left to right based on their basal transcription in p53 +/+ cells over p53 −/− cells. Green indicates genes whose basal transcription is greater than twofold in p53 +/+ cells, red indicates lesser than twofold. Grey dots display the transcription of the same genes in Nutlin-treated p53 +/+ cells. (B) Heatmap displaying relative transcriptional activity of direct p53 target genes identified by GRO-seq relative to control p53 −/− cells. Genes are sorted based on their transcription in control p53 +/+ cells. (C) Genome browser views of representative genes whose basal transcription is higher (GDF15) or lower (PTP4A1) in the presence of MDM2-bound p53. See Figure 3—figure supplement 1A for matching RNAPII ChIP data. (D) Q-RT-PCR measurements of genes whose basal transcription was found to be 2x higher (green) or lower (red) in the presence of MDM2-bound p53. (E) ChIP assays show binding of p53 and MDM2 to the p53REs in the CDKN1A and PTP4A1 gene loci (−2283 bp and +1789 relative to TSS, respectively), prior to inhibition of the p53-MDM2 interaction by Nutlin. Nutlin treatment leads to increased p53 signals with the DO-1 antibody recognizing the p53 TAD1, concurrently with a decrease in MDM2 signals. MDM2 ChIP was performed in SJSA cells carrying a MDM2 gene amplification F. Oncomine gene expression analysis of 598 cancer cell lines of varied p53 status shows that CDKN1A, DDB2 and GDF15 are more highly expressed in wild type p53 cell lines, whereas GJB5 is more highly expressed in mutant p53 cell lines. The ranking position of these genes is also indicated.