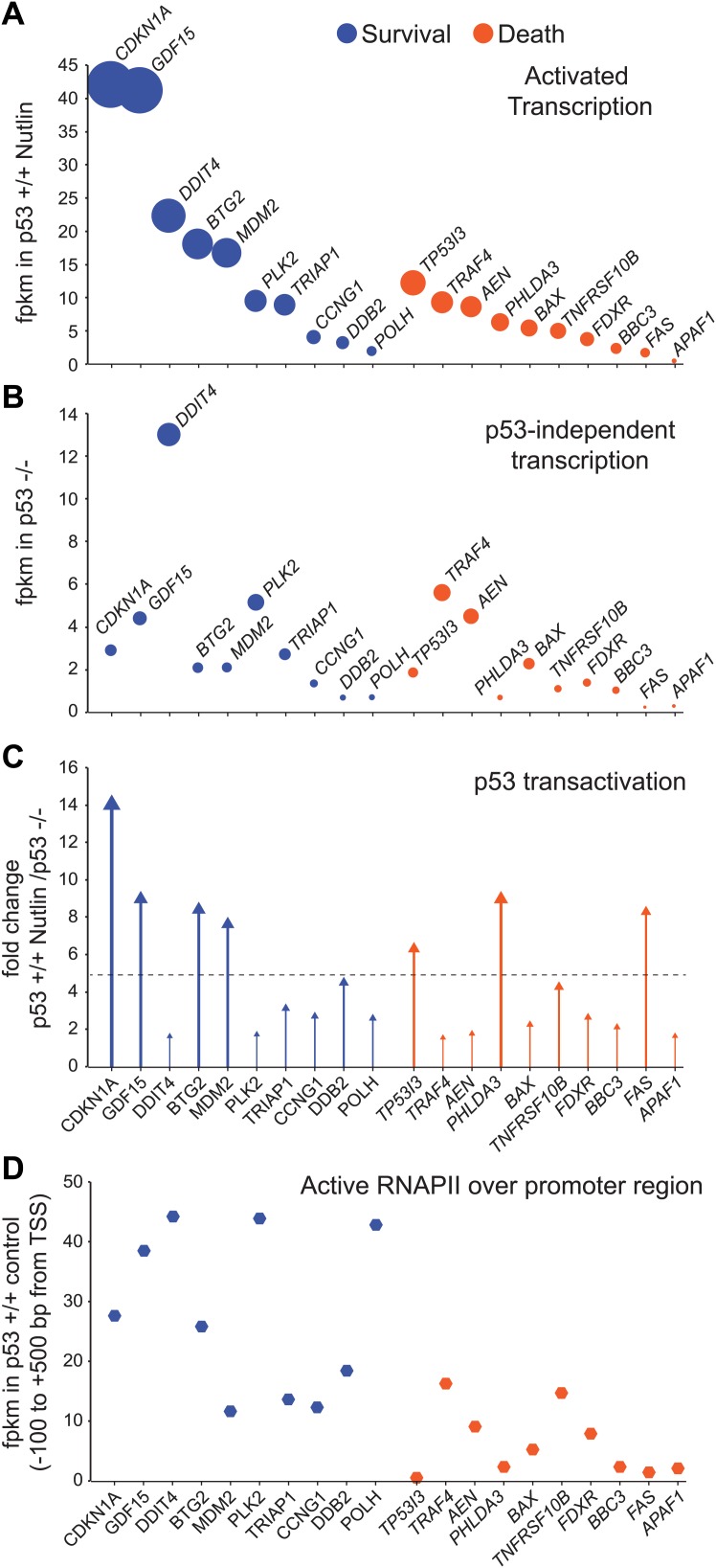
Figure 4. GRO-seq analysis of key survival and death genes within the p53 network.
(A) The 10 most transcribed pro-survival and pro-apoptotic genes identified by GRO-seq ranked by decreasing transcriptional output in Nutlin-treated p53 +/+ cells. The surface of the bubbles represents the GRO-seq signal output relative to the CDKN1A locus. (B) Transcriptional output of same genes shown in A in p53 −/− cells. (C) Fold change analysis showing the overall effect of p53 on the transcription of its survival and apoptotic targets. (D) Survival genes within the p53 network tend to carry more proximally bound, transcriptionally engaged RNAPII over their promoter regions than apoptotic genes.