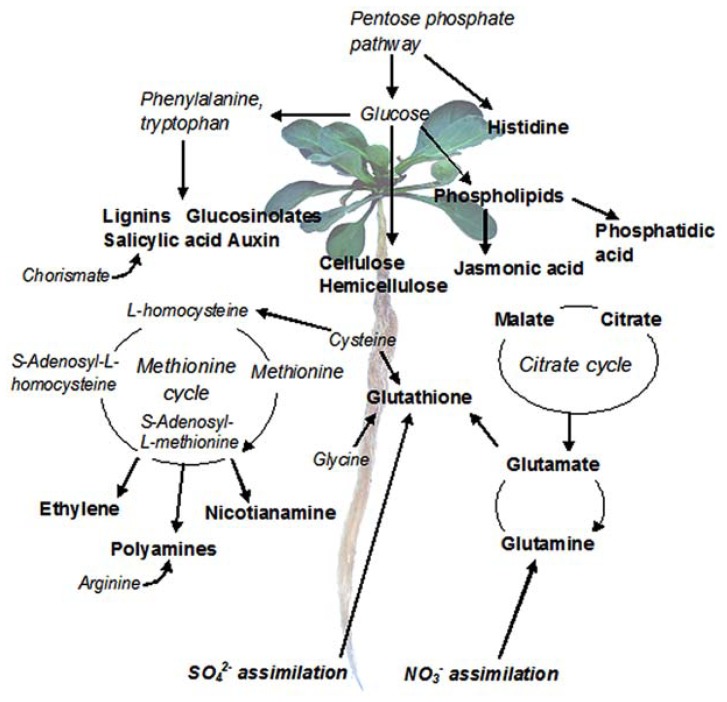
FIGURE 2.
The most active metabolic pathways in N. caerulescens roots. The data are based on SOLiD RNA-Seq analysis of three biological replicates of three N. caerulescens accessions: La Calamine, Ganges, and Monte Prinzera. The read counts were normalized by the size of the libraries and the length of the genes, assuming an equal length for orthologous genes in A. thaliana and N. caerulescens, and the most highly expressed genes (expression value > 1000) were subjected to a more detailed analysis. The genes were manually linked to metabolic pathways in KEGG (Kyoto Encyclopedia of Genes and Genomes).