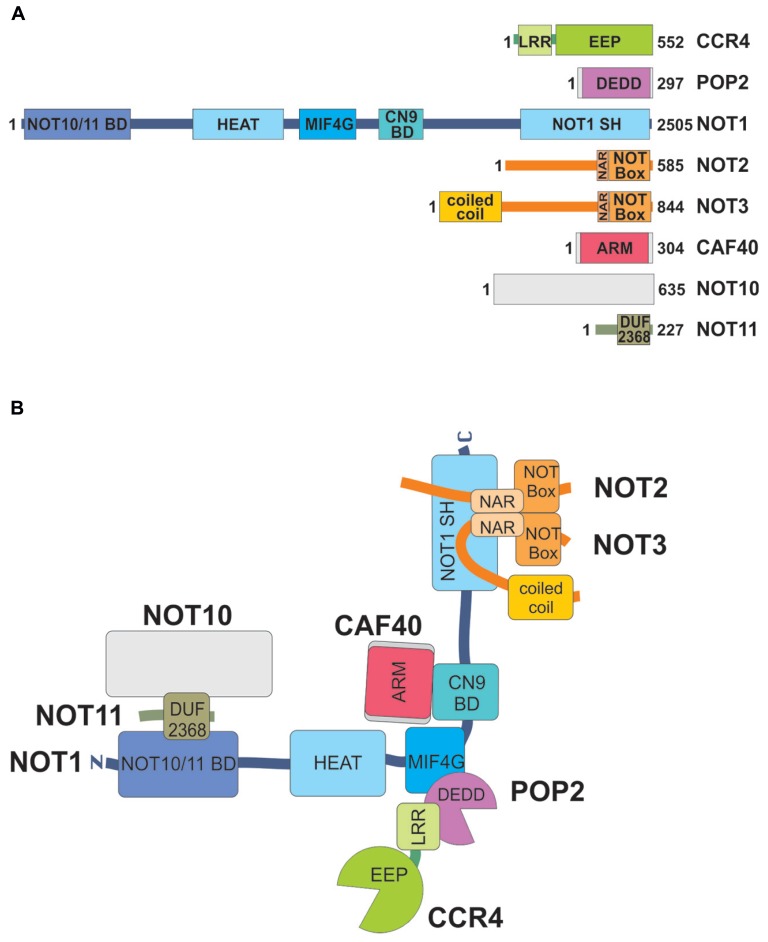
FIGURE 1.
Subunits of the Drosophila CCR4–NOT complex, their domains and interactions. (A) Domain structures of subunits. The size of each polypeptide (in amino acids) is indicated on the right. Note that this can vary due to alternative splicing. Each large rectangle corresponds to a structured domain. CCR4 contains a leucine-rich repeat (LRR) and an exonuclease-endonuclease-phosphatase (EEP) domain. POP2 consists of a single nuclease domain of the DEDD class. NOT1 contains a NOT10/NOT11 binding domain at its N-terminus, a series of HEAT repeats, a middle-of-4G (MIF4G) domain, and a CAF40/CNOT9 binding domain (CN9BD) in the middle. The C-terminus is formed by a conserved NOT1 superfamily homology (NOT1 SH) domain. The MIF4G, and the NOT1 SH domains are also composed of HEAT repeats. Both NOT2 and NOT3 contain a C-terminal NOT box preceded by a NOT1 anchor region (NAR), NOT3 also has a predicted N-terminal coiled–coil domain. CAF40 consists of armadillo (ARM) repeats. NOT10 has no known or predicted domain, and NOT11 contains a domain of unknown function. (B) Interactions between the subunits of the CCR4–NOT complex. Domains are indicated with the same color code as in (A). The orientation of the coiled–coil domain of NOT3 is arbitrary. Interactions shown are based on experimental data for the Drosophila complex and comparison to the yeast and mammalian complexes (Bawankar et al., 2013). The L-shape indicated for the complex is based on cryo EM images (Nasertorabi et al., 2011).