Abstract
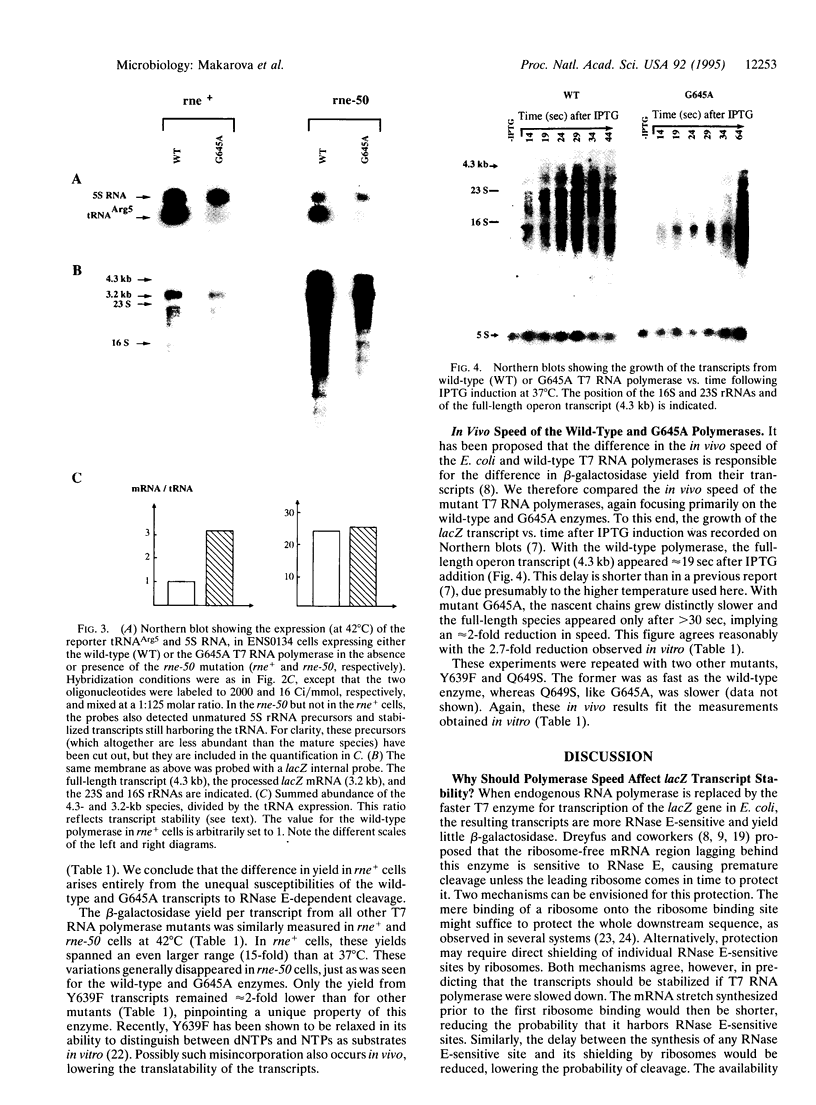
When in Escherichia coli the host RNA polymerase is replaced by the 8-fold faster bacteriophage T7 enzyme for transcription of the lacZ gene, the beta-galactosidase yield per transcript drops as a result of transcript destabilization. We have measured the beta-galactosidase yield per transcript from T7 RNA polymerase mutants that exhibit a reduced elongation speed in vitro. Aside from very slow mutants that were not sufficiently processive to transcribe the lacZ gene, the lower the polymerase speed, the higher the beta-galactosidase yield per transcript. In particular, a mutant which was 2.7-fold slower than the wild-type enzyme yielded 3.4- to 4.6-fold more beta-galactosidase per transcript. These differences in yield vanished in the presence of the rne-50 mutation and therefore reflect the unequal sensitivity of the transcripts to RNase E. We propose that the instability of the T7 RNA polymerase transcripts stems from the unmasking of an RNase E-sensitive site(s) between the polymerase and the leading ribosome: the faster the polymerase, the longer the lag between the synthesis of this site(s) and its shielding by ribosomes, and the lower the transcript stability.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. M., Chamberlin M. J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990 May 5;213(1):79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Bechhofer D. H., Zen K. H. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):5803–5811. doi: 10.1128/jb.171.11.5803-5811.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
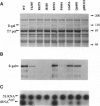
- Bonner G., Lafer E. M., Sousa R. Characterization of a set of T7 RNA polymerase active site mutants. J Biol Chem. 1994 Oct 7;269(40):25120–25128. [PubMed] [Google Scholar]
- Bonner G., Patra D., Lafer E. M., Sousa R. Mutations in T7 RNA polymerase that support the proposal for a common polymerase active site structure. EMBO J. 1992 Oct;11(10):3767–3775. doi: 10.1002/j.1460-2075.1992.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. Y., Kan M. C., Lin-Chao S. RNAII transcribed by IPTG-induced T7 RNA polymerase is non-functional as a replication primer for ColE1-type plasmids in Escherichia coli. Nucleic Acids Res. 1995 May 25;23(10):1691–1695. doi: 10.1093/nar/23.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier-Miller M., Jacques N., Raibaud O., Dreyfus M. Transcription of single-copy hybrid lacZ genes by T7 RNA polymerase in Escherichia coli: mRNA synthesis and degradation can be uncoupled from translation. Nucleic Acids Res. 1990 Oct 11;18(19):5787–5792. doi: 10.1093/nar/18.19.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., Schlessinger D. Coupling of rates of transcription, translation, and messenger ribonucleic acid degradation in streptomycin-dependent mutants of Escherichia coli. J Bacteriol. 1976 Jan;125(1):84–93. doi: 10.1128/jb.125.1.84-93.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995 Jul 3;14(13):3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994 Nov 10;372(6502):193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- Iost I., Guillerez J., Dreyfus M. Bacteriophage T7 RNA polymerase travels far ahead of ribosomes in vivo. J Bacteriol. 1992 Jan;174(2):619–622. doi: 10.1128/jb.174.2.619-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J Mol Biol. 1971 Sep 28;60(3):453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- Lewicki B. T., Margus T., Remme J., Nierhaus K. H. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993 Jun 5;231(3):581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
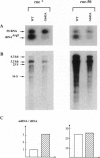
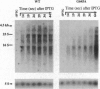
- Lopez P. J., Iost I., Dreyfus M. The use of a tRNA as a transcriptional reporter: the T7 late promoter is extremely efficient in Escherichia coli but its transcripts are poorly expressed. Nucleic Acids Res. 1994 Apr 11;22(7):1186–1193. doi: 10.1093/nar/22.7.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. K., Kolesnikow T., Rayner J. C., Simons E. L., Yim H., Simons R. W. Control of translation by mRNA secondary structure: the importance of the kinetics of structure formation. Mol Microbiol. 1994 Dec;14(5):1033–1047. doi: 10.1111/j.1365-2958.1994.tb01337.x. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. L., Jr, Hamkalo B. A., Thomas C. A., Jr Visualization of bacterial genes in action. Science. 1970 Jul 24;169(3943):392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Murakawa G. J., Kwan C., Yamashita J., Nierlich D. P. Transcription and decay of the lac messenger: role of an intergenic terminator. J Bacteriol. 1991 Jan;173(1):28–36. doi: 10.1128/jb.173.1.28-36.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979 Apr 15;129(3):343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Hearst J. E. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J Biol Chem. 1988 Jan 5;263(1):527–534. [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995 Sep 15;14(18):4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Stent G. S. Genetic transcription. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):181–197. doi: 10.1098/rspb.1966.0022. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Vogel U., Sørensen M., Pedersen S., Jensen K. F., Kilstrup M. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol Microbiol. 1992 Aug;6(15):2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Yarchuk O., Jacques N., Guillerez J., Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1992 Aug 5;226(3):581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]