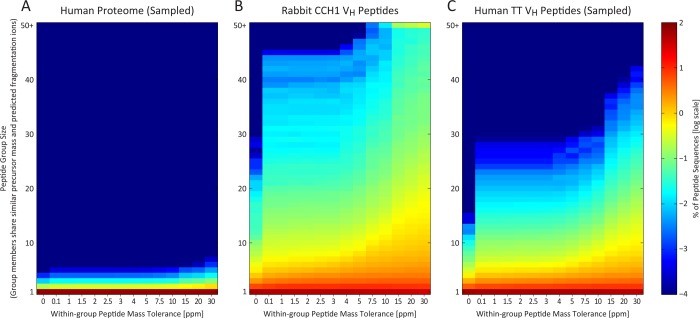
Figure 2.
In contrast to the proteome in general, antibody peptide sequences resemble each other in both mass and expected fragmentation patterns. The peptide sequence search space is thus strongly dependent on mass accuracy, as seen by plotting the extent of theoretical peptide-spectral match ambiguity, for (A) human proteome peptide sequences, (B) rabbit CCH antibody VH peptides, and (C) human tetanus toxoid antibody VH peptides. Reducing precursor mass tolerance thus more strongly affects the potential for false identifications in VH peptides than for a typical proteome. Here, an in silico digest of the rabbit CCH VH antibody sequences generated 505 790 unique peptide sequences (constrained to fully tryptic peptides of ≥8 amino acids, ≤6000 Da theoretical mass, and ≤2 missed cleavages). Each peptide sequence contributes to a y-axis bin defined by the self-inclusive count of all theoretical peptides within a specified mass tolerance (x-axis) and sharing at least 60% predicted fragmentation ion similarity. For comparison, the human proteome (A) and human TT VH (C) sequence databases were processed likewise and subsampled to include the same number of peptide sequences as (B). The intersequence similarity evident in the antibody sets is negligible in this size-matched human proteome control.