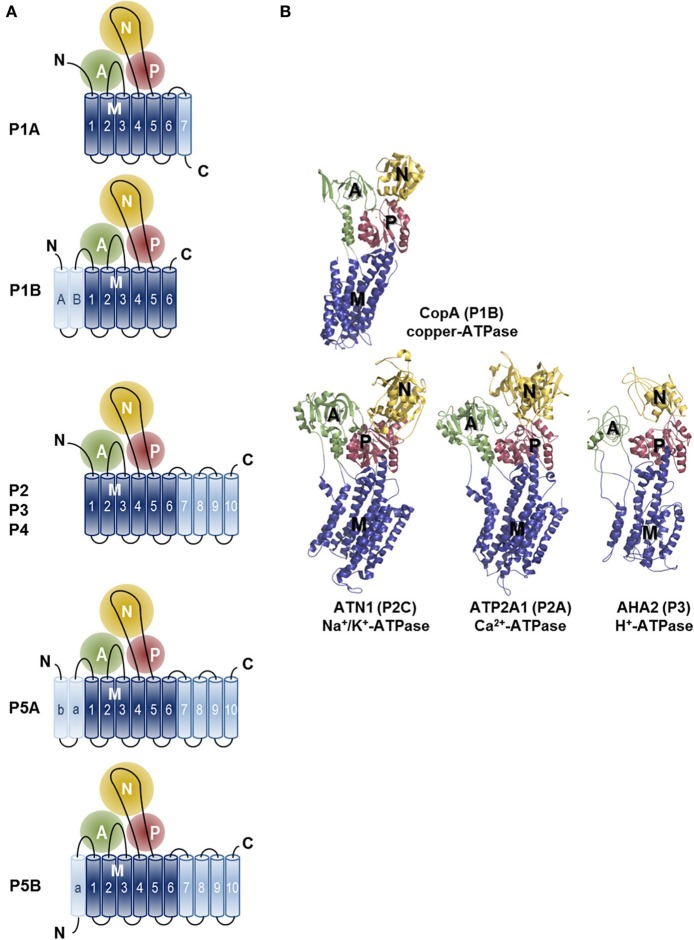
Figure 3.
Topology and architecture of the catalytic subunits of P-type ATPases. (A) Planar topology models of the five classes of P-type ATPases (P1-P5). Nucleotide-binding domains (N, yellow), actuator domains (A, green) and phosphorylation domains (P, red) are indicated. The 6 TM helices (Polymeropoulos et al., 1997; Spillantini et al., 1997; Jankovic, 2008; Lees et al., 2009; Auluck et al., 2010; Tolleson and Fang, 2013) form the core segment of the membrane (M) domain of all P-type ATPases, which is depicted in dark blue, whereas additional N- and C-terminal helices are shown in light blue. Of note, there is one exception for the P2 ATPases, a splice variant of ATP2A2, SERCA2b, harbors an 11th TM helix at the C-terminus (not shown, Vandecaetsbeek et al., 2009). (B) Resolved P-type ATPase crystal structures, known up until now. The Legionella pneumophila CopA copper-ATPase (PDB 3RFU), a P1B-type ATPase; the rabbit P2A-type ATPase ATP2A1 (SERCA1a, PDB 2AGV), the Squalus acanthias Na+/K+-ATPase α-subunit ATN1 (PDB 3A3Y), a member of the P2C group and the Arabidopsis thaliana proton pump AHA2 (PBD 3B8C) of the P3-type ATPases. N-, A-, P- and M-domains are indicated with similar colors as in the planar models. Note that the obligatory subunits of the P1A, P2C and P4 are not shown.