Abstract
Systemic lupus erythematosus (SLE) disease is an autoimmune disease of unknown aetiology that affects predominantly women of child bearing age. Since previous studies, including ours, have demonstrated that CD4+ T cells and B cells from SLE patients are defective in their ability to methylate their DNA upon antigen stimulation, the aim of this study was to investigate whether DNA demethylation affects the transcription of HRES-1 in B cells. HRES-1 is the prototype of Human Endogenous Retrovirus (HERV) overexpressed in SLE. We have observed that SLE B cells were characterized by their incapacity to methylate the HRES-1 promoter, both in unstimulated and in anti-IgM stimulated B cells. In turn, HRES-1/p28 expression was increased in SLE B cells after B cell receptor engagement, but not in controls. In SLE B cells the Erk/DNMT1 pathway was defective. In addition, blocking the autocrine-loop of IL-6 in SLE B cells with an anti-IL-6 receptor monoclonal antibody restores DNA methylation and control of HRES-1/p28 expression became effective. As a consequence, a better understanding of HERV dysregulation in SLE reinforces our comprehension of the disease and opens new therapeutic perspectives.
Keywords: B cells, DNA methylation, Erk, HRES-1, IL-6, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) disease is an auto-immune disease with a large spectrum of clinical and immunological manifestations that affects predominantly women of child-bearing age [1]. The aetiology of SLE is multifactorial including genetic, epigenetic, environmental and hormonal factors. DNA methylation, the main epigenetic process, is abnormal in CD4+ T cells and B cells from SLE patients (Reviewed in [2]) leading to the overexpression of DNA methylation sensitive genes such as Human Endogenous Retroviruses (HERV). HERV detection is increased in patients with autoimmune diseases including SLE [3,4], Sjögren’s syndrome [5,6], and multiple sclerosis [7]. HERV are not only suspected to be markers of autoimmune diseases they are also suspected of contributing to the development of autoimmune diseases through different mechanisms.
In this way, HERV proteins have been demonstrated to impact immune regulation by producing cross-reactive autoantibodies (Ab) by molecular mimicry [8], or by expressing viral genes with super-antigen properties [9]. Another possibility is that HERV genetic elements interfere with neighboring immune related genes in an abnormal way as described in B cells with the cell surface receptor CD5 [10,11].
HERV were first discovered in the late 1960s, and their involvement since then has been described in all vertebrates [12]. In primates, up to 8% of the genome contains HERV elements, which are 4-fold more than the coding sequences and 7-fold more when considering the number of genes [13]. Phylogenetic analysis has highlighted up to 30 distinct groups ranging from one to many thousand copy numbers [14,15]. HERV chromosomal duplication started over one hundred million years ago and the process is still ongoing. Duplication occurs randomly in a copy-and-paste fashion via an intermediate RNA step. Mutations and deletions are frequently observed explaining why full length HERV (~10 kb) are exceptions [16]. HERV are composed of two long terminal repeats (LTR) in the 5′ and 3′ ends and between them three genes gag, pol, and env may be present.
In SLE, the HERV prototype is HRES-1, which is located at position 1q42 within chromosome 1 [17]. The link between HRES-1 and SLE started with the observation that anti-HRES-1/p28 gag autoAb are detected in up to 50% of SLE patients in contrast to less than 5% in the healthy control groups [18], and that within SLE patients HRES-1/p28 gag protein cross-reacts with the autoantigen U1-snRNP [8]. Next, the contribution of HRES-1 in the aetiology of SLE was reinforced by the observation that several HRES-1 haplotypes were linked with SLE [19,20] and that over-expression of the reverse transcript HRES1-/Rab4 in CD4+ T cells affects TCR signaling [21]. Recently we have suspected that DNA methylation represses HRES-1/p28 expression in healthy control B cells and that this effect can be reversed in the presence of IL-6 [10]. The aim of the present study was to explore whether HRES-1/p28 expression in SLE B cells is related to DNA methylation.
Materials and methods
B-lymphocyte isolation
Peripheral blood was collected from 6 patients with inactive SLE (SLE disease activity index <5) and 6 healthy controls (HC). All patients fulfilled the American College of Rheumatology criteria for SLE [22,23]. Informed consent was obtained from the patients before collecting blood, and the Institutional Review Board at Brest University Medical School approved the study protocol.
Using centrifugation on Ficoll-Hypaque (PAA Laboratories, Linz, Austria), peripheral blood mononuclear cells (PBMC) were isolated and then B cells were negatively purified using the EasySep™ enrichment Kit (Stemcell Technologies Inc., Vancouver, Canada). All purified cells were >98% CD19+.
Cell culture
B cells were suspended in RPMI-1640 media supplemented with 10% heat-inactivated fetal calf sera, 2 mM L-glutamine, 200 U/ml penicillin and 100 μg/ml streptomycin (Lonza Inc., Allendale, NJ). B cells were seeded at 5 × 105 cells per well, and incubated 24 h with 1 μg/ml of anti-IgM Ab-coated Sepharose beads (Bio-Rad, Hercules, CA) and 10 U/ml IL-2, in the presence or absence of 40 ng/ml anti-IL-6R Ab (R&D Systems, Minneapolis, MN), or 100 ng/ml rhIL-6 (ImmunoTools, Friesoythe, Germany). Inhibition of DNMTs was achieved by incubating the cells 24 h with 20 μM of 5-azacytidine (5-aza) or with 50 μM of the ras signal blocker PD98059 (Sigma-Aldrich, St Louis, MO).
Methylation-specific PCR
Genomic DNA was purified using QIAmp 96 DNA blood kit (Qiagen, Carlsbad, CA) and 100 ng template DNA was distributed into three aliquots. The DNA concentration and the 260:280 nm absorbance ratios were calculated using a nanodrop 2000c spectrophotometer (Thermoscientific Nanodrop Technologies, Wilmington, DE). The first aliquot was undigested and used as a positive control. The second aliquot was digested with 20 U of the methylation-insensitive restriction enzyme MspI for 3 h at 37 °C (New England Biolabs, Beverly, MA). The third aliquot was digested with 20 U of HpaII for 3 h at 37 °C. This assay is based on the inability of HpaII restriction enzymes to digest a methylated 5′-CCmGG-3′ site. Next, the PCR was conducted using primers positioned downstream 5′-GCATATGCACTG GGAAAGGT-3′ and upstream 5′-CCGCCTTTTCAAGTTTC CTC-3′ of the unique HpaII/MspI CCGG site present within the HRES-1 promoter (Figure 1A). The PCR protocol included an initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 sec, annealing at 56 °C for 1 min, and primer extension at 72 °C for 1 min; PCR cycles were followed by final extension at 72 °C for 10 min. The PCR products were separated on an agarose gel and visualized with 0.5 μg/ml ethidium bromide.
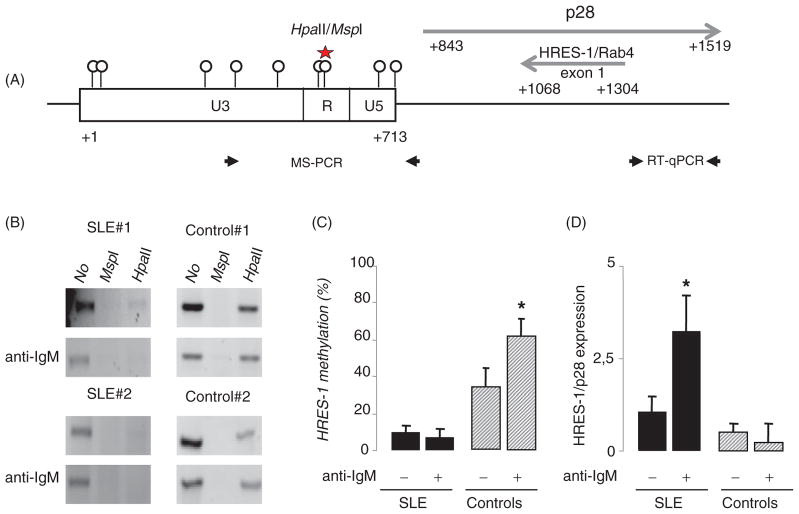
Figure 1.
The HRES-1 promoter region is demethylated in B cells from SLE patients and DNA demethylation modifies HRES-1/p28 expression. A: Schematic representation of the organization of HRES-1 on chromosome 1q42 region (GenBank X16660). Circles identify the 9 CpG motifs within the 5′ U3-R-U5 LTR region of HRES-1. Of note, the 3′ LTR region of HRES-1 is lacking. The HpaII/MspI motif present in the 5′U3-R-U5 LTR region is represented (star) as well as HRES-1/p28 and HRES-1/Rab4 transcript positions, and primers used in the MS-PCR and quantitative RT-PCR (RT-qPCR). B: Analysis of HRES-1 promoter methylation by amplification of genomic DNA digested with the methylation sensitive HpaII, or with the methylation insensitive MspI enzyme. The 502 bp HRES-1 amplicon contains one HpaII/MspI site. C: Histograms representing HRES-1 promoter methylation status in 6 SLE patients and 6 healthy controls, the percentage of methylation was quantified by calculating the ratio of HpaII-digested to undigested bands. D: Quantitative RT-PCR results presenting as histograms revealing that a 24-h stimulation of B cells with an anti-IgM increases HRES-1/p28 transcription in B cells from SLE patients, but not from healthy controls. The symbol * represented p<0.05.
mRNA extraction and quantitative RT-PCR
Total mRNA was extracted using the RNAble method (Eurobio, Les-Ullis, France), and cDNA synthesized by reverse transcription in 20 μl volume with Superscript™ II RNase H-RT (Invitrogen Life Sciences, Carlsbad, CA). Quantitative RT-PCR was carried out in 20-μl mixtures containing 50 ng template cDNA, 1X Sybr® Green PCR Master mix (Applied Biosystems, Foster City, CA), and 500 nM of each primer, HRES-1/p28 sense primer 5′-GGAA GAGGAGATGGGCTACG-3′ and HRES-1/p28 reverse primer 5′-CAGGGAAATCGGGACTCAG-3′. All assays included a negative control, which consisted of the reaction mixture with no template, and a positive control, which consisted of the mixture with 18S rRNA. Comparison of cycle thresholds was completed with the 2−ΔΔct method using 18S as an internal control.
Flow cytometry
Intracellular phosphorylated mitogen-activated protein kinase/extracellular-regulated kinase (MAPK/pErk1/2) staining (Dako, Carpinteria, CA) and DNA methyl transferase 1 (DNMT1) staining (Abcam, Cambridge, UK) were performed, respectively, after permeabilization using the cytofix/cytoperm intracellular staining kit (BD Biosciences, San Jose, CA) or permeabilization using 70% methanol in purified B-cells stimulated or not with 1 μg/ml of anti-IgM Ab (Bio-Rad). Next, anti-pErk1/2 binding was revealed after 10 min anti-IgM stimulation and DNMT1 binding after 24 h anti-IgM stimulation using a F(ab)′2 goat FITC anti-murine IgG (Jackson Immunosearch, West Grove, MO). Results were expressed in mean fluorescence intensity (MFI).
Statistical analysis
The results are expressed in arithmetic means with standard deviations (SD). Data were compared using the Mann–Whitney U-test for unpaired data. Significance was assessed as p<0.05.
Results
Impaired HRES-1 promoter methylation in SLE B cells explains HRES-1/p28 overexpression in anti-IgM stimulated B cells
Analysis of the 713 pb 5′ U3-R-U5 promoter sequence of HRES-1 (Genbank X16660) reveals 9 CpG motifs including a HpaII/MspI 5′-CCGG-3′ motif at position +559 (Figure 1A). Next, in order to test HRES-1 promoter methylation status, a MS-PCR was developed using specific primers flanking the HpaII/MspI motif. The PCR was conducted using either undigested DNA (Figure 1B lane 1, referred to as 100% methylation), DNA digested with the methylation-insensitive enzyme MspI (lane 2, 0% methylation), and DNA digested with the methylation-sensitive enzyme HpaII (lane 3).
The HpaII MS-PCR analysis of HRES-1 promoter methylation status in unstimulated and anti-IgM stimulated B cells from 6 SLE patients and 6 healthy controls (HC) revealed that HRES-1 promoter was hypomethylated in SLE B cells both when analyzing unstimulated B cells (HRES-1 methylation status: 10.2% ± 2.3% in SLE B cells versus 35.1% ± 10.9% in HC B cells, p<0.05) and when analyzing anti-IgM stimulated B cells (6.9% ± 5.3% in SLE B cells versus 62.8% ± 12.1% in HC B cells, p<0.01) (Figure 1C). To determine the functional consequences of DNA methylation changes in HRES-1 promoter (Figure 1D), we performed a gene expression analysis. HRES-1/p28 mRNA levels were not different when considering unstimulated B cells between SLE patients and HC. After B cell receptor (BCR) engagement, HRES-1/p28 mRNA expression was increased 3.2-fold in SLE B cells and repressed 2.1-fold in HC B cells. These results indicate that induction of HRES-1/p28 following BCR engagement is regulated at the epigenetic level.
The Erk/DNMT1 pathway is defective in anti-IgM stimulated SLE B cells
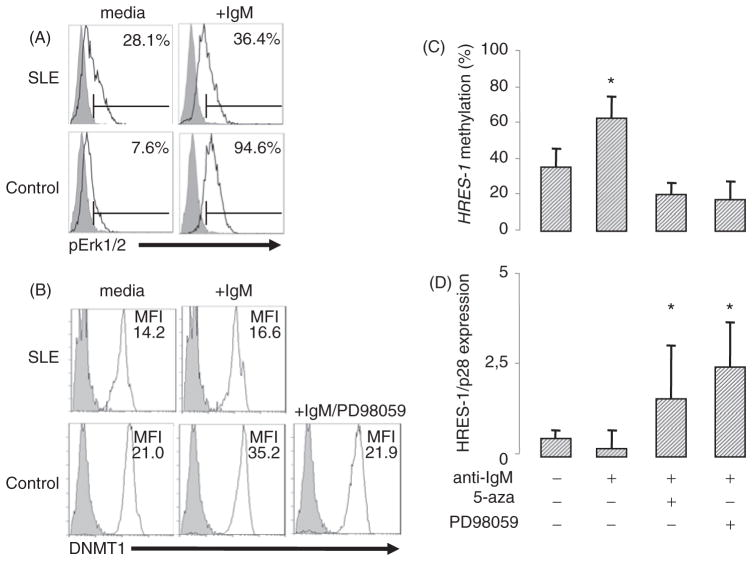
As described in SLE T cells [24,25], the incapacity of SLE B cells to methylate their DNA may result from a defective Erk/DNMT1 pathway. To test this hypothesis, SLE B cells and HC B cells were stimulated with an anti-IgM Ab and the intracellular phosphorylation status of the MAPK Erk was evaluated after 10 min by flow cytometry. As shown in Figure 2(A) in a representative experiment, the MAPK Erk was strongly phosphorylated in HC B cells after BCR engagement (pErk1/2: 7.5 ± 3.8% versus 73.5 ± 19.9 with anti-IgM, p<0.05), but not in SLE B cells (pErk1/2: 24.1 ± 4.4% versus 27.5 ± 5.6 with anti-IgM, non significant). Next, the role of the MAPK Erk pathway on DNMT1 induction was examined. In HC B cells, but not in SLE B cells, DNMT1 expression was induced after 24 h anti-IgM stimulation (MFI DNMT1: 22.7 ± 2.7% versus 32.6 ± 6.5 with anti-IgM in HC, p<0.05) and this induction was repressed in the presence of the MEK/Erk inhibitor PD98059 (Figure 2B).
Figure 2.
The Erk/DNMT pathway is defective in SLE B cells. A–B: Cytoplasmic staining of pErk (A) and DNMT1 (B) in permeabilized B cells stimulated with anti-IgM or not for 10 min (pErk) and 24 h (DNMT1). Representative flow cytometric profiles are represented. C: HRES-1 promoter analysis by MS-PCR reveals that the Erk/DNMT1 pathway was involved in HRES-1 promoter methylation when control B cells were BCR stimulated with an anti-IgM Ab. Inhibition of pErk and DNMTs was respectively achieved by incubating the cells 24 h with 50 μM of the ras signal blocker PD98059, and with 20 μM of 5-azacytidine (5-aza). D: Quantitative PCR measurement of HRES-1/P28 mRNA levels. The symbol * represented p<0.05.
To confirm the contribution of the MAPK Erk/DNMT1 pathway to the control of DNA methylation, anti-IgM stimulation was repeated in HC B cells either in the presence of PD98059, or in the presence of the DNMT1 inhibitor 5-azacytidine. As shown in Figure 2(C and D), Erk and DNMT1 inhibitors affect the anti-IgM-dependent HRES-1 promoter DNA methylation process and as a consequence this inhibition contribute to the enhancement of HRES-1/p28 mRNA expression.
The role of IL-6 in demetlylation and HRES-1/p28 expression
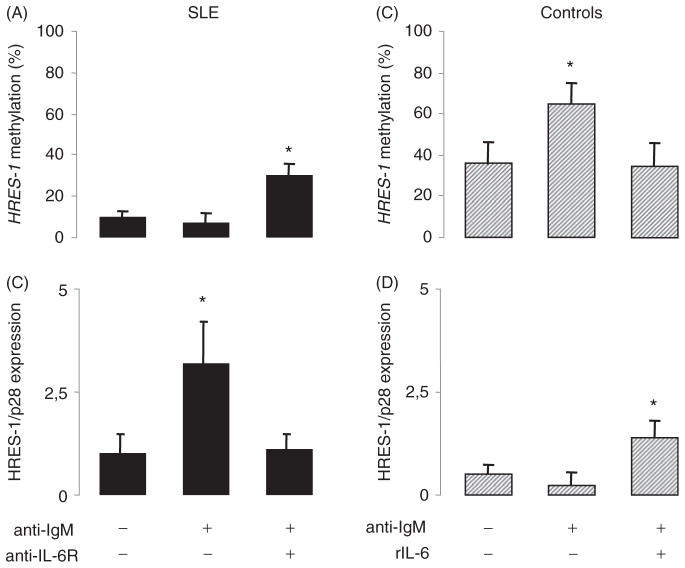
Finally, based on the control of DNMT1 by IL-6 in B cells [10], and to test the effect of the SLE B cell autocrine loop of IL-6 [26] on HRES-1/p28 expression, a blocking anti-IL6 receptor was used in anti-IgM stimulated SLE B cells. As reported in Figure 3(A–B), blocking IL-6 in SLE B cells restores anti-IgM-dependent HRES-1 promoter methylation, which represses HRES-1/p28 expression. To further highlight the importance of IL-6 in the control of DNA methylation, HC B cells were anti-IgM-stimulated in the presence of rIL-6 (Figure 3C–D). The addition of rIL-6 affects the ability of the anti-IgM stimulation to methylate the HRES-1 promoter, thus permitting HRES-1/p28 expression in HC B cells.
Figure 3.
The effect of IL-6 on HRES-1/P28 expression and HRES-1 promoter methylation status. A/C: HRES-1 promoter analysis by MS-PCR in SLE B cells (A) and healthy control B cells (C) after BCR engagement using an anti-IgM Ab. Based on our previous observation that anti-IgM stimulated SLE B cells, but not the controls, express high amounts of IL-6 [26], the influence of the autocrine loop of IL-6 was tested by adding 40 ng/ml of a blocking anti-IL-6 receptor Ab in SLE B, or by adding 100 ng/ml of rIL-6 in healthy control B cells. B/D: Quantitative PCR measurement of HRES-1/P28.
Discussion
In this report we have observed that (i) DNA methylation is impaired in resting and anti-IgM stimulated SLE B cells, that (ii) HRES-1/p28 expression is regulated by DNA methylation through the Erk/DNMT1 pathway, and that (iii) this defect is reversible when blocking the autocrine loop of IL-6.
Results generated from our previous studies and others indicated that demethylation of DNA can be related to the inhibition or lack of DNMTs, to an increase of demethylating activity, or from a combination of both mechanisms [10,27]. No association between DNMT1 polymorphism and SLE was observed [28]. In SLE, several DNA demethylating external factors such as hydralazine used as primary drug for treating hypertension [24], ultraviolet B irradiation [29], and gender [30] have been associated with SLE susceptibility and flare severity in genetically predisposed individuals. In addition, internal DNA demethylating factors may be also suspected such as cell cycle arrest [10], viral infection [31], and in response to cellular communication. We have recently observed that DNA demethylation in salivary glands from patients with primary Sjögren’s syndrome is related to the infiltrating B cells [32]. Last but not least, several microRNA have been reported in SLE to target DNMT1 directly or indirectly [33,34].
In CD4+, CD8+ and CD19+ B cells isolated from SLE patients DNA demethylation is associated with disease activity, leucopenia, lymphopenia and UVB irradiation with differences observed according to the lymphocyte subset [35,36]. The genome-wide methylation analysis has been recently explored, from these studies several genes relevant to SLE including type I interferon genes were highlighted [37–40]. Interestingly, most of the DNA methylation dysregulated genes were previously identified by genome-wide association studies (GWAS) [41].
As a consequence, altered methylation influences T cells in SLE causing up-regulation of co-stimulating molecules (CD70, CD40 ligand), integrins (LFA-1) and cytokines (IL-1, IL-10, IL-13) [42]. In SLE, we proposed that B cell dysfunctions [43,44] are also altered by DNA demethylation [45] and the contribution of DNA demethylated B cells in the autoimmune process has been demonstrated in mice [46].
Aside from CpG-rich sequences within the genes, CpG are present within “junk DNA” to control DNA repetitive transposons and retroelements, respectively 2.8% and 41.5% of the human genome [47]. Retroelements can further be divided in three groups: long interspersed nuclear elements (LINEs, 20.1% of the human genome), short interspersed nuclear elements (ALU, 10.6%; MIR 2.5%), and HERVs (8.3%). In lymphocytes from SLE patients, the DNA demethylation process is selective, since ALU sequences are not concerned [37,48], while LINE-1 sequences are demethylated and among HERV elements the HERV-E family is particularly sensitive to DNA methylation [3,49].
Consistent with this observation is the finding that the HERV-E element HERV-CD5 is deregulated at the epigenetic level in SLE B cells [10]. HRES-1 and HERV-CD5 share several caracteristics (i) chromosomal integration occurs at the stage of old word primate divergence [16], (ii) they are both class1 HERVs and related to the C-type retroviruses [50], (iii) they are silenced by DNA methylation, and (iiii) they interfere with immune genes, respectively, CD4 and CD5 [51,52]. Divergences between HRES-1 and HERV-CD5 are related (i) to the HERV family sub-class, (ii) to the absence of HRES-1 3′LTR, and (iii) to the B cell specificity for HERV-CD5 [53].
The proposed involvement of IL-6 in promoting auto-reactivity is supported by in vitro and in vivo studies leading to the development of an anti-IL6 receptor blocking mAb, tocilizumab, which is effective in SLE [54]. In patients treated with tocilizumab, IL-6 repression is associated with the decrease of lymphocyte activation and the normalization of the B and T cell subsets [55]. The precise mechanism, by which IL-6 interferes with lymphocyte homeostasis, remains to be elucidated. According to our report, part of the effect may be related to the action of IL-6 on DNA methylation. Interestingly, mutations of DNMT3b as observed in immuno-deficiency, centromere instability and facial anomalies (ICF) syndrome lead to major dysregulation of lymphogenesis [56,57].
In conclusion, the finding that DNA methylation controls HERV transcription is relevant for understanding mechanisms that control autoreactivity. In addition, the data provide further arguments to consider in the development of epigenetic based therapies in SLE.
Acknowledgments
Thanks are due to Geneviève Michel and Simone Forest for their help with typing of the paper. We are also grateful to the “Association Française du Gougerot Sjögren et des Syndromes Secs” and the “Association Lupus France”.
Footnotes
Declaration of interest
The authors declare no competing financial interests This paper was supported in part by grants AI048079 and AI072648 from the National Institutes of Health. YT is funded by the Islamic development bank merit scholarship program. The authors alone are responsible for the content and writing of the paper.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Renaudineau Y, Youinou P. Epigenetics and auto-immunity, with special emphasis on methylation. Keio J Med. 2011;60:10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 3.Ogasawara H, Naito T, Kaneko H, et al. Quantitative analyses of messenger RNA of human endogenous retrovirus in patients with systemic lupus erythematosus. J Rheumatol. 2001;28:533–538. [PubMed] [Google Scholar]
- 4.Okada M, Ogasawara H, Kaneko H, et al. Role of DNA methylation in transcription of human endogenous retrovirus in the pathogenesis of systemic lupus erythematosus. J Rheumatol. 2002;29:1678–1682. [PubMed] [Google Scholar]
- 5.Le Dantec C, Varin MM, Brooks WH, et al. Epigenetics and Sjogren’s syndrome. Curr Pharm Biotechnol. 2012;13:2046–2053. doi: 10.2174/138920112802273326. [DOI] [PubMed] [Google Scholar]
- 6.Thabet Y, Canas F, Ghedira I, et al. Altered patterns of epigenetic changes in systemic lupus erythematosus and auto-antibody production: is there a link? J Autoimmun. 2012;39:154–160. doi: 10.1016/j.jaut.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Montojo M, Dominguez-Mozo M, Arias-Leal A, et al. The DNA copy number of human endogenous retrovirus-W (MSRV-type) is increased in multiple sclerosis patients and is influenced by gender and disease severity. PLoS One. 2013;8:e53623. doi: 10.1371/journal.pone.0053623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perl A, Colombo E, Dai H, et al. Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum. 1995;38:1660–1671. doi: 10.1002/art.1780381119. [DOI] [PubMed] [Google Scholar]
- 9.Sicat J, Sutkowski N, Huber BT. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol. 2005;32:1821–1831. [PubMed] [Google Scholar]
- 10.Garaud S, Le Dantec C, Jousse-Joulin S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. 2009;182:5623–5632. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- 11.Garaud S, Le Dantec C, Berthou C, et al. Selection of the alternative exon 1 from the cd5 gene down-regulates membrane level of the protein in B lymphocytes. J Immunol. 2008;181:2010–2018. doi: 10.4049/jimmunol.181.3.2010. [DOI] [PubMed] [Google Scholar]
- 12.Brooks WH, Le Dantec C, Pers JO, et al. Epigenetics and autoimmunity. J Autoimmun. 2010;34:J207–219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Solyom S, Kazazian HH., Jr Mobile elements in the human genome: implications for disease. Genome Med. 2012;4:12. doi: 10.1186/gm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blomberg J, Benachenhou F, Blikstad V, et al. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renaudineau Y, Vallet S, Le Dantec C, et al. Characterization of the human CD5 endogenous retrovirus-E in B lymphocytes. Genes Immun. 2005;6:663–671. doi: 10.1038/sj.gene.6364253. [DOI] [PubMed] [Google Scholar]
- 17.Perl A, Rosenblatt JD, Chen IS, et al. Detection and cloning of new HTLV-related endogenous sequences in man. Nucl Acids Res. 1989;17:6841–6854. doi: 10.1093/nar/17.17.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banki K, Maceda J, Hurley E, et al. Human T-cell lymphotropic virus (HTLV)-related endogenous sequence, HRES-1, encodes a 28-kDa protein: a possible autoantigen for HTLV-I gag-reactive autoantibodies. Proc Natl Acad Sci USA. 1992;89:1939–1943. doi: 10.1073/pnas.89.5.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magistrelli C, Samoilova E, Agarwal RK, et al. Polymorphic genotypes of the HRES-1 human endogenous retro-virus locus correlate with systemic lupus erythematosus and autoreactivity. Immunogenetics. 1999;49:829–834. doi: 10.1007/s002510050561. [DOI] [PubMed] [Google Scholar]
- 20.Pullmann R, Jr, Bonilla E, Phillips PE, et al. Haplotypes of the HRES-1 endogenous retrovirus are associated with development and disease manifestations of systemic lupus erythematosus. Arthritis Rheum. 2008;58:532–540. doi: 10.1002/art.23161. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez DR, Telarico T, Bonilla E, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol. 2009;182:2063–2073. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petri M, Orbai AM, Alarcon GS, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–2686. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 24.Deng C, Lu Q, Zhang Z, et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 25.Deng C, Kaplan MJ, Yang J, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Hillion S, Garaud S, Devauchelle V, et al. Interleukin-6 is responsible for aberrant B-cell receptor-mediated regulation of RAG expression in systemic lupus erythematosus. Immunology. 2007;122:371–380. doi: 10.1111/j.1365-2567.2007.02649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balada E, Ordi-Ros J, Serrano-Acedo S, et al. Transcript overexpression of the MBD2 and MBD4 genes in CD4+ T cells from systemic lupus erythematosus patients. J Leukoc Biol. 2007;81:1609–1616. doi: 10.1189/jlb.0107064. [DOI] [PubMed] [Google Scholar]
- 28.Park BL, Kim LH, Shin HD, et al. Association analyses of DNA methyltransferase-1 (DNMT1) polymorphisms with systemic lupus erythematosus. J Hum Genet. 2004;49:642–646. doi: 10.1007/s10038-004-0192-x. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Zhao M, Yin H, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62:1438–1447. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 30.Sawalha AH, Wang L, Nadig A, et al. Sex-specific differences in the relationship between genetic susceptibility, T cell DNA demethylation and lupus flare severity. J Autoimmun. 2012;38:J216–J222. doi: 10.1016/j.jaut.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernando H, Shannon-Lowe C, Islam AB, et al. The B cell transcription program mediates hypomethylation and overexpression of key genes in Epstein–Barr virus-associated proliferative conversion. Genome Biol. 2013;14:R3. doi: 10.1186/gb-2013-14-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thabet Y, Le Dantec C, Ghedira I, et al. Epigenetic dysregulation in salivary glands from patients with primary Sjogren’s syndrome may be ascribed to infiltrating B cells. J Autoimmun. 2013;41:175–181. doi: 10.1016/j.jaut.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyl-transferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 34.Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4(+) T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2010;63:1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 35.Wang GS, Zhang M, Li XP, et al. Ultraviolet B exposure of peripheral blood mononuclear cells of patients with systemic lupus erythematosus inhibits DNA methylation. Lupus. 2009;18:1037–1044. doi: 10.1177/0961203309106181. [DOI] [PubMed] [Google Scholar]
- 36.Nakkuntod J, Sukkapan P, Avihingsanon Y, et al. DNA methylation of human endogenous retrovirus in systemic lupus erythematosus. J Hum Genet. 2013;58:241–249. doi: 10.1038/jhg.2013.6. [DOI] [PubMed] [Google Scholar]
- 37.Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeffries MA, Dozmorov M, Tang Y, et al. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coit P, Jeffries M, Altorok N, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furukawa H, Oka S, Matsui T, et al. Genome, epigenome and transcriptome analyses of a pair of monozygotic twins discordant for systemic lupus erythematosus. Hum Immunol. 2013;74:170–175. doi: 10.1016/j.humimm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: recent progress from GWAS. J Autoimmun. 2013;41:25–33. doi: 10.1016/j.jaut.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Lu Q, Renaudineau Y, Cha S, et al. Epigenetics in autoimmune disorders: highlights of the 10th Sjogren’s syndrome symposium. Autoimmun Rev. 2010;9:627–630. doi: 10.1016/j.autrev.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Taher TE, Muhammad HA, Bariller E, et al. B-Lymphocyte signalling abnormalities and lupus immunopathology. Int Rev Immunol. 2013;32:428–444. doi: 10.3109/08830185.2013.788648. [DOI] [PubMed] [Google Scholar]
- 44.Taher TE, Muhammad HA, Rahim A, et al. Aberrant B-lymphocyte responses in lupus: inherent or induced and potential therapeutic targets. Eur J Clin Invest. 2013;43:866–880. doi: 10.1111/eci.12111. [DOI] [PubMed] [Google Scholar]
- 45.Garaud S, Youinou P, Renaudineau Y. DNA methylation and B-cell autoreactivity. Adv Exp Med Biol. 2011;711:50–60. doi: 10.1007/978-1-4419-8216-2_5. [DOI] [PubMed] [Google Scholar]
- 46.Mazari L, Ouarzane M, Zouali M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc Natl Acad Sci USA. 2007;104:6317–6322. doi: 10.1073/pnas.0610434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta. 2011;412:1457–1461. doi: 10.1016/j.cca.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Piotrowski PC, Duriagin S, Jagodzinski PP. Expression of human endogenous retrovirus clone 4-1 may correlate with blood plasma concentration of anti-U1 RNP and anti-Sm nuclear antibodies. Clin Rheumatol. 2005;24:620–624. doi: 10.1007/s10067-005-1123-8. [DOI] [PubMed] [Google Scholar]
- 50.Adelman MK, Yocum DE, Marchalomis JJ. Endogenous retroviruses are etiological agents in systemic lupus erythematosus. In: Shoenfeld Y, Rose NR, editors. Infection and Autoimmunity. Elsevier; The Netherlands: 2004. pp. 271–288. [Google Scholar]
- 51.Renaudineau Y, Hillion S, Saraux A, et al. An alternative exon 1 of the CD5 gene regulates CD5 expression in human B lymphocytes. Blood. 2005;106:2781–2789. doi: 10.1182/blood-2005-02-0597. [DOI] [PubMed] [Google Scholar]
- 52.Nagy G, Ward J, Mosser DD, et al. Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem. 2006;281:34574–34591. doi: 10.1074/jbc.M606301200. [DOI] [PubMed] [Google Scholar]
- 53.Garaud S, Morva A, Lemoine S, et al. CD5 promotes IL-10 production in chronic lymphocytic leukemia B cells through STAT3 and NFAT2 activation. J Immunol. 2011;186:4835–4844. doi: 10.4049/jimmunol.1003050. [DOI] [PubMed] [Google Scholar]
- 54.Illei GG, Shirota Y, Yarboro CH, et al. Tocilizumab in systemic lupus erythematosus: data on safety, preliminary efficacy, and impact on circulating plasma cells from an open-label phase I dosage-escalation study. Arthritis Rheum. 2010;62:542–552. doi: 10.1002/art.27221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirota Y, Yarboro C, Fischer R, et al. Impact of anti-interleukin-6 receptor blockade on circulating T and B cell subsets in patients with systemic lupus erythematosus. Ann Rheum Dis. 2013;72:118–128. doi: 10.1136/annrheumdis-2012-201310. [DOI] [PubMed] [Google Scholar]
- 56.Blanco-Betancourt CE, Moncla A, Milili M, et al. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood. 2004;103:2683–2690. doi: 10.1182/blood-2003-08-2632. [DOI] [PubMed] [Google Scholar]
- 57.Heyn H, Vidal E, Sayols S, et al. Whole-genome bisulfite DNA sequencing of a DNMT3B mutant patient. Epigenetics. 2012;7:542–550. doi: 10.4161/epi.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]