Significance
The ubiquitin–proteasome system (UPS) is required for Drosophila class IV dendritic arborization neuron dendrite pruning. We found that mutants in the ubiquitylation machinery, the ubiquitin-dependent chaperone Valosin-Containing Protein (VCP), and the 19S regulatory particle of the proteasome—but not the 20S core particle—showed defects in pruning gene expression and mislocalization or overexpression of specific mRNA-binding proteins. In the case of VCP inhibition, we were able to detect a specific change in the splicing pattern of a pruning gene that likely contributes to pruning defects. A link between VCP and mRNA-binding proteins had been observed in the context of human neurodegenerative disease. This study implicates a specific function of VCP and ubiquitin in mRNA metabolism.
Abstract
The dendritic arbors of the larval Drosophila peripheral class IV dendritic arborization neurons degenerate during metamorphosis in an ecdysone-dependent manner. This process—also known as dendrite pruning—depends on the ubiquitin–proteasome system (UPS), but the specific processes regulated by the UPS during pruning have been largely elusive. Here, we show that mutation or inhibition of Valosin-Containing Protein (VCP), a ubiquitin-dependent ATPase whose human homolog is linked to neurodegenerative disease, leads to specific defects in mRNA metabolism and that this role of VCP is linked to dendrite pruning. Specifically, we find that VCP inhibition causes an altered splicing pattern of the large pruning gene molecule interacting with CasL and mislocalization of the Drosophila homolog of the human RNA-binding protein TAR–DNA-binding protein of 43 kilo-Dalton (TDP-43). Our data suggest that VCP inactivation might lead to specific gain-of-function of TDP-43 and other RNA-binding proteins. A similar combination of defects is also seen in a mutant in the ubiquitin-conjugating enzyme ubcD1 and a mutant in the 19S regulatory particle of the proteasome, but not in a 20S proteasome mutant. Thus, our results highlight a proteolysis-independent function of the UPS during class IV dendritic arborization neuron dendrite pruning and link the UPS to the control of mRNA metabolism.
To achieve specific connections during development, neurons need to refine their axonal and dendritic arbors. This often involves the elimination of neuronal processes by regulated retraction or degeneration, processes known collectively as pruning (1). In the fruit fly Drosophila melanogaster, large-scale neuronal remodeling and pruning occur during metamorphosis. For example, the peripheral class IV dendritic arborization (da) neurons specifically prune their extensive larval dendritic arbors (2, 3), whereas another class of da neurons, the class III da neurons, undergo ecdysone- and caspase-dependent cell death (3). Class IV da neuron dendrite pruning requires the steroid hormone ecdysone (2, 3) and its target gene SOX14, encoding an HMG box transcription factor (4). Class IV da neuron dendrites are first severed proximally from the soma by the action of enzymes like Katanin-p60L and Mical that sever microtubules and actin cables, respectively (4, 5). Later, caspases are required for the fragmentation and phagocytic engulfment of the severed dendrite remnants (6, 7). Another signaling cascade known to be required for pruning is the ubiquitin–proteasome system (UPS) (2, 7, 8). Covalent modification with the small protein ubiquitin occurs by a thioester cascade involving the ubiquitin-activating enzyme Uba1 (E1), and subsequent transfer to ubiquitin-conjugating enzymes (E2s) and the specificity-determining E3 enzymes (9). Ubiquitylation of a protein usually leads to the degradation of the modified protein by the proteasome, a large cylindrical protease that consists of two large subunits, the 19S regulatory particle and the proteolytic 20S core particle (10). Several basal components of the ubiquitylation cascade—Uba1 and the E2 enzyme ubcD1—as well as several components of the 19S subunit of the proteasome have been shown to be required for pruning (2, 7, 8), as well as the ATPase associated with diverse cellular activities (AAA) ATPase Valosin-Containing Protein (VCP) (CDC48 in yeast, p97 in vertebrates, also known as TER94 in Drosophila) (11), which acts as a chaperone for ubiquitylated proteins (12). Interestingly, autosomal dominant mutations in the human VCP gene cause hereditary forms of ubiquitin-positive frontotemporal dementia (FTLD-U) (13) and amyotrophic lateral sclerosis (ALS) (14). A hallmark of these diseases is the occurrence of both cytosolic and nuclear ubiquitin-positive neuronal aggregates that often contain the RNA-binding protein TAR–DNA-binding protein of 43 kilo-Dalton (TDP-43) (15). We previously proposed that ubcD1 and VCP promote the activation of caspases during dendrite pruning via degradation of the caspase inhibitor DIAP1 (7, 11). However, mutation of ubcD1 or VCP inhibit the severing of class IV da neuron dendrites from the cell body (7, 11), whereas in caspase mutants, dendrites are still severed from the cell body, but clearance of the severed fragments is affected (6, 16). This indicates that the UPS must have additional, as yet unidentified, functions during pruning.
Here, we further investigated the role of UPS mutants in dendrite pruning. We show that vcp mutation leads to a specific defect in ecdysone-dependent gene expression, as VCP is required for the functional expression and splicing of the large ecdysone target gene molecule interacting with CasL (MICAL). Concomitantly, we observe mislocalization of Drosophila TDP-43 and up-regulation of other RNA-binding proteins, and genetic evidence suggests that these alterations contribute to the observed pruning defects in VCP mutants. Defects in MICAL expression and TDP-43 localization are also induced by mutations in ubcD1 and in the 19S regulatory particle of the proteasome, but not by a mutation in the 20S core particle, despite the fact that proteasomal proteolysis is required for dendrite pruning, indicating the requirement for multiple UPS pathways during class IV da neuron dendrite pruning.
Results and Discussion
VCP Mutant Pruning Phenotypes Are Linked to Ecdysone Signaling.
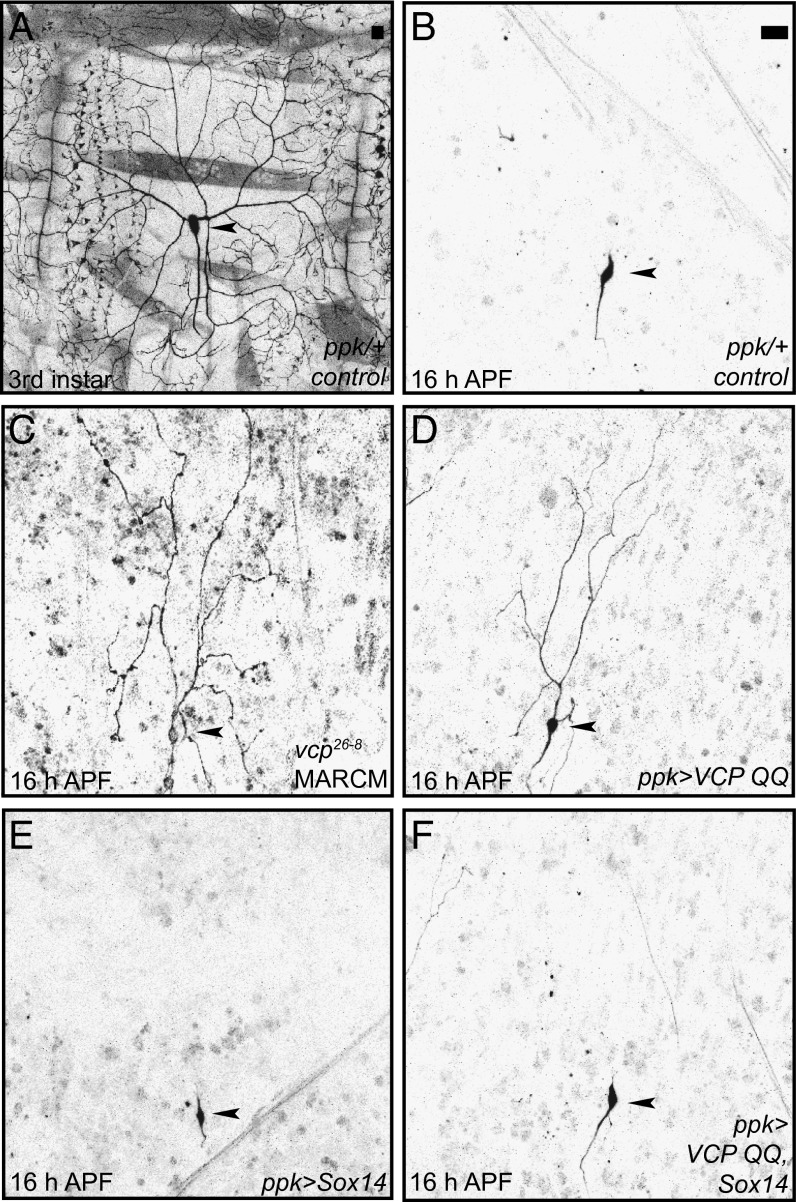
Class IV da neurons have long and branched dendrites at the third instar larval stage (Fig. 1A). In wild-type animals, these dendrites are completely pruned at 16–18 h after puparium formation (h APF) (Fig. 1B). We next generated VCP mutant class IV da neurons by the Mosaic Analysis with a Repressible Cell Marker (MARCM) technique for clonal analysis (17). Mutant vcp26-8 class IV da neurons displayed strong pruning defects and retained long dendrites at 16 h APF (Fig. 1C). As previously shown (11), expression of an ATPase-deficient dominant-negative VCP protein (VCP QQ) under the class IV da neuron-specific driver ppk-GAL4 recapitulated the pruning phenotype and also led to the retention of long and branched dendrites at 16 h APF (Fig. 1D). We had previously shown that VCP inhibition also causes defects in class III da neuron apoptosis (11). This combination of defects in both pruning and apoptosis is reminiscent of the phenotypes caused by defects in ecdysone-dependent gene expression (3, 4). Indeed, we noted that overexpression of the transcription factor Sox14, which induces pruning genes, led to a nearly complete suppression of the pruning phenotype caused by VCP QQ (Fig. 1 D–F). This genetic interaction suggested that VCP might be required for the expression of one or several ecdysone target genes during pruning.
Fig. 1.
VCP requirement for dendrite pruning is linked to ecdysone signaling. (A) Morphology of the dorsal class IV da neuron ddaC at the third instar larval stage. (B) ddaC morphology at 16 h APF. All larval dendrites are pruned (0/38 with dendrites attached to soma). (C) vcp26-8 mutant ddaC neurons at 16 h APF show severe defects in dendrite pruning (8/8 attached to soma, P < 0.005). (D) Expression of dominant-negative VCP QQ in class IV da neurons inhibits dendrite pruning at 16 h APF (18/30 with dendrites attached to soma, P < 0.005). (E and F) Overexpression of the ecdysone pathway transcription factor Sox14 suppresses the pruning defects induced by VCP QQ. (E) Sox14 overexpression does not affect dendrite pruning at 16 h APF (0/34). (F) Sox14 overexpression suppresses dendrite pruning defects induced by VCP QQ (0/50). Statistical comparison of attached versus severed dendrites was with Fisher’s exact test. (Scale bars in A and B, 20 μm.) Arrowheads denote the positions of class IV da neuron cell bodies.
VCP Regulates Mical Expression.
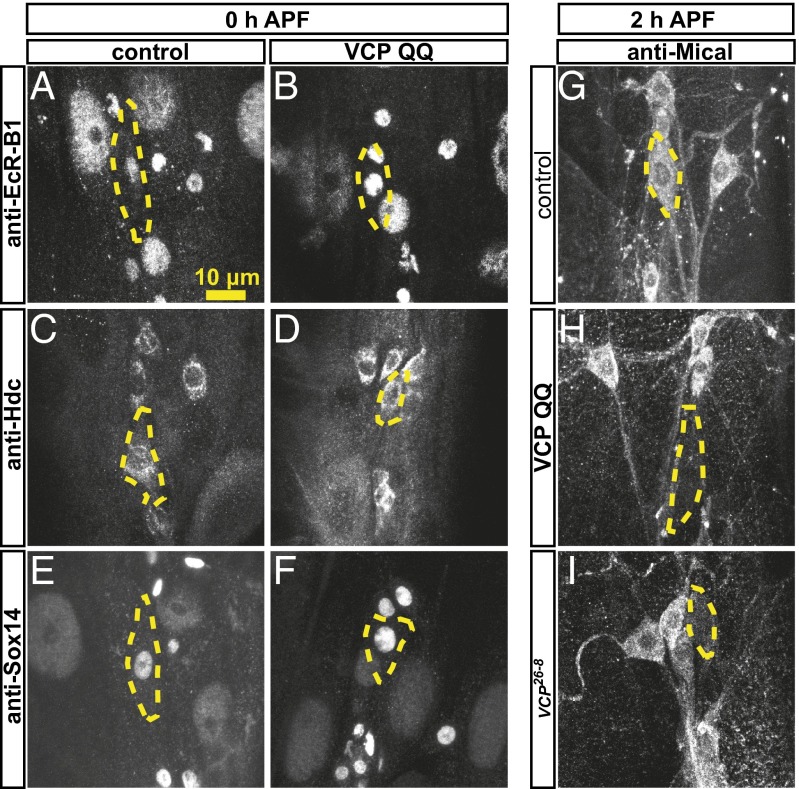
How could VCP be linked to Sox14? The suppression of the vcp mutant phenotype by Sox14 overexpression could be achieved in one of several ways. Sox14 could be epistatic to VCP—that is, VCP could be required for functional Sox14 expression—and this effect would be mitigated by Sox14 overexpression. However, VCP could also be required for the expression of one or several Sox14 target genes, and enhanced Sox14 expression could overcome this requirement either via enhanced induction of one or several particular targets or via enhanced induction of other pruning genes, in which case Sox14 would be a bypass suppressor of VCP QQ. To distinguish between these possibilities, we next assessed the effects of VCP inhibition on the expression of known genes in the ecdysone cascade required for pruning in class IV da neurons. Class IV da neuron pruning is governed by the Ecdysone Receptor B1 (EcR-B1) isoform (2, 3), which in turn directly activates the transcription of Sox14 (4) and Headcase (Hdc), a pruning factor of unkown function (18). Sox14, on the other hand, activates the transcription of the MICAL gene encoding an actin-severing enzyme (4). In immunostaining experiments, VCP QQ did not affect the expression of EcR-B1 (Fig. 2 A and B), Sox14 (Fig. 2 C and D), or Hdc (Fig. 2 E and F) at the onset of the pupal phase. However, the expression of Mical was selectively abrogated in class IV da neurons expressing VCP QQ (Fig. 2 G and H), or in vcp26-8 class IV da neuron MARCM clones (Fig. 2I). These data indicated that VCP might affect dendrite pruning by regulating the expression of the Sox14 target gene Mical, indicating that Sox14 might act as a bypass suppressor of VCP QQ.
Fig. 2.
VCP inhibition leads to specific down-regulation of the ecdysone target and pruning factor Mical at the onset of the pupal phase. Expression of ecdysone pathway components in the class IV da neuron ddaC upon VCP inhibition was assessed by immunofluorescence with specific antibodies. (A and B) Expression of EcR-B1 at the white pupal stage (0 h APF) in a control class IV da neuron (A) or a class IV da neuron expressing VCP QQ. (C and D) Expression of Hdc at the white pupal stage in (C) control and in (D) neurons expressing VCP QQ. (E and F) Expression of Sox14 at the white pupal stage: (E) control and (F) with VCP QQ. (G–I) Expression of Mical at 2 h APF: (G) control, (H) with VCP QQ, and (I) in a vcp26-8 mutant MARCM ddaC neuron. (Scale bar, 20 μm.) Class IV da neuron cell bodies are outlined by dashed lines.
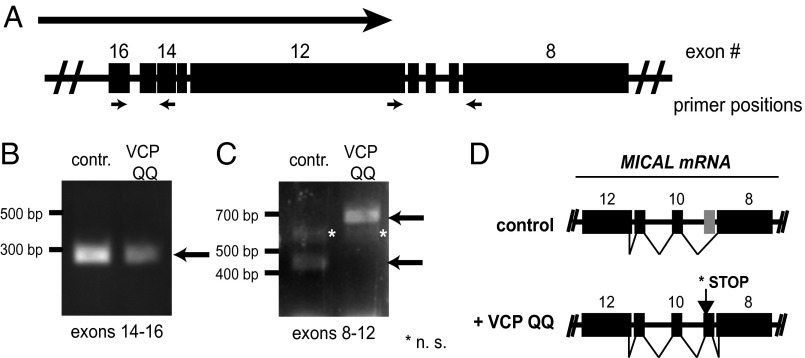
How could VCP inhibition suppress Mical expression? To answer this question, we next assessed if Mical mRNA could still be detected in class IV da neurons expressing VCP QQ. To this end, we used enzymatic tissue digestion and FACS sorting (19) to isolate class IV da neurons from early pupae (1–5 h APF). We then extracted total RNA from the isolated neurons and assessed the presence of Mical mRNA expression by RT-PCR, using control samples or samples from animals expressing VCP QQ under ppk-GAL4. The Mical gene is large (∼40 kb) and spans multiple exons that are transcribed to yield a ∼15 kb mRNA. To detect Mical cDNA, we used primer pairs spanning several exons for two different regions of Mical mRNA, exons 14–16 and exons 8–12 (Fig. 3A). [MICAL is on the (–) strand, but the exon numbering denoted by flybase.org follows the direction of the (+) strand. Therefore, exons 14–16 are upstream of exons 8–12, and the latter are closer to the 3′ end of MICAL mRNA.] MICAL mRNA was detectable upon VCP inhibition in these extracts with a primer pair spanning exons 14–16. The second primer pair spanning exons 8–12 also detected MICAL mRNA in both samples (Fig. 3C), but the RT-PCR product from the VCP QQ-expressing class IV da neurons had a larger molecular weight. Sequencing of the PCR products indicated that MICAL mRNA from VCP QQ-expressing class IV da neurons contained exon 11, which was not present in Mical mRNA from the control sample (Fig. 3D). Exon 11 is absent from all predicted MICAL splice isoforms except for a weakly supported isoform designated “Mical-RM” (www.flybase.org). It introduces a stop codon into MICAL mRNA that would lead to the truncation of the C-terminal 1,611 amino acids from Mical protein. This portion of Mical protein contains several predicted protein interaction domains such as a proline-rich region, a coiled-coil region with similarity to Ezrin/Radixin/Moesin (ERM) domains, and a C-terminal PDZ-binding motif, and is required for the interaction between Mical and PlexinA (20). In addition, the truncated region contains the epitope for the antibody used (20) in the immunofluorescence experiments, thus explaining the observed lack of Mical expression upon VCP inhibition. Given that a mutant of Mical with a smaller C-terminal truncation (compared with the one induced by VCP inhibition) was not sufficient to rescue the class IV da neuron dendrite pruning defect of mical mutants (4), disruption of VCP function likely results in expression of a truncated Mical protein without pruning activity. Taken together, these data suggest that the observed defect in MICAL mRNA splicing contributes significantly to the pruning defects of VCP mutants.
Fig. 3.
MICAL mRNA is misspliced upon VCP inhibition. Total RNA was isolated from control class IV da neurons or class IV da neurons expressing VCP QQ at 1–5 h APF and analyzed by RT-PCR. (A) Schematic representation of part of the MICAL gene. The large arrow indicates the direction of transcription and the small arrows the positions of RT-PCR primers. The exon numbering is as on flybase.org. (B and C) RT-PCR reactions from control or VCP QQ-expressing samples: (B) MICAL exons 14–16 and (C) MICAL exons 8–12. (D) Schematic representation of change in MICAL splicing upon VCP inhibition. The stop codon induced by VCP QQ would delete 1,612 amino acids from the Mical protein compared with the full-length protein.
VCP Regulates RNA-Binding Proteins in Class IV da Neurons.
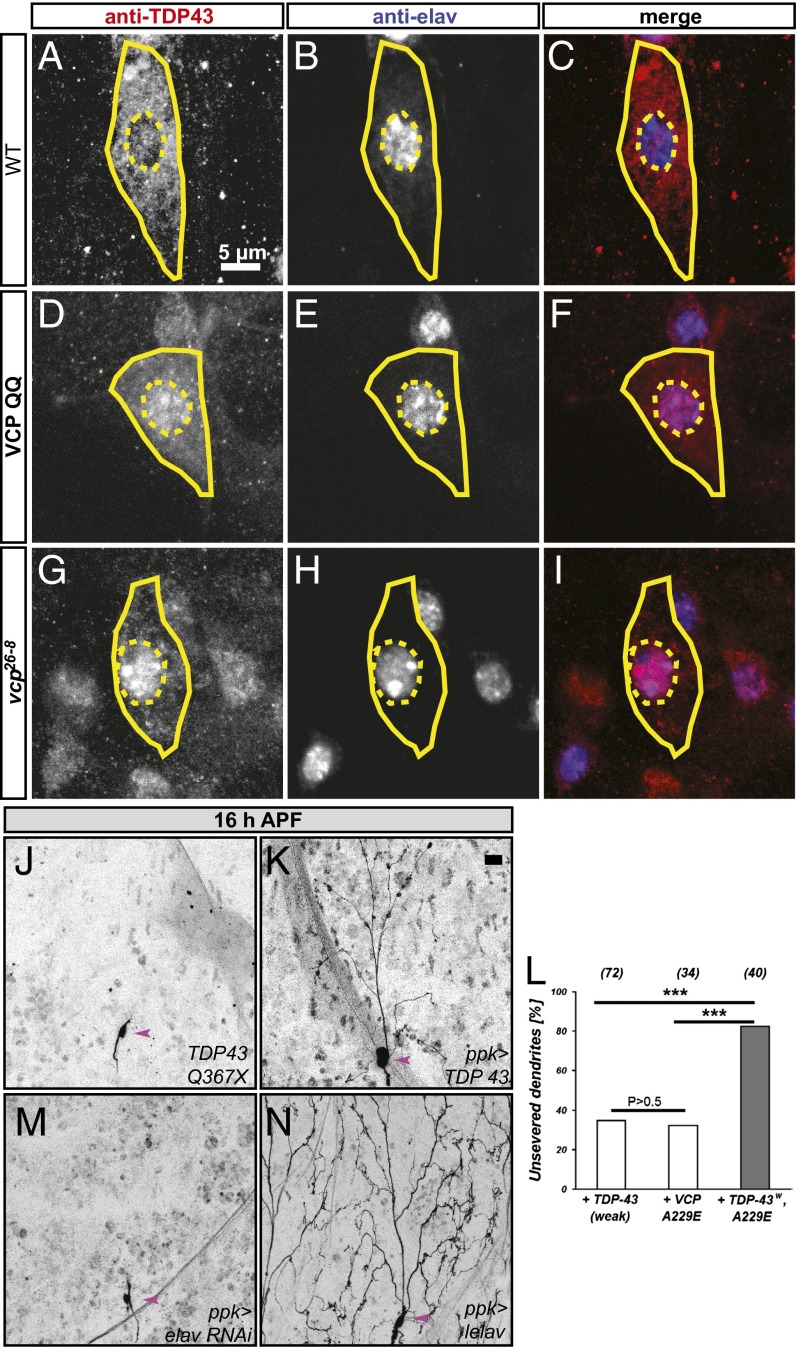
How is VCP linked to alternative splicing of MICAL mRNA? A plausible mechanism for the control of an alternative splicing event would be the modulation of specific (pre)mRNA-binding proteins. VCP has recently been linked to several RNA-binding proteins: human autosomal dominant VCP mutations cause frontotemporal dementia or ALS with inclusion bodies that contain aggregated human TDP-43 (14, 15); a genetic screen in Drosophila identified the RNA-binding proteins Drosophila TDP-43, HRP48, and x16 as weak genetic interactors of the dominant effects of VCP disease mutants (21); and HuR (a human homolog of the neuronal Drosophila RNA-binding protein elav) was recently shown to bind human VCP (22). Of these, TDP-43 and also elav have been linked to alternative splicing in various model systems, including Drosophila (23–25). We therefore used available specific antibodies to assess the levels and distribution of Drosophila TDP-43 (hereafter referred to as TDP-43) and elav. TDP-43 has previously been shown to localize to the nucleus in Drosophila motoneurons and mushroom body Kenyon cells (26). Surprisingly, TDP-43 was largely localized to the cytoplasm in class IV da neurons, where it was enriched in a punctate pattern around the nucleus, with only a small fraction also detectable in the nucleus (Fig. 4 A and C), a localization pattern that could be reproduced with transgenic N-terminally HA-tagged TDP-43 (Fig. S1). elav is a known nuclear marker for Drosophila neurons; in class IV da neurons, it was somewhat enriched in nuclear punctae (Fig. 4 B and C). We next assessed the effects of VCP inhibition on these two RNA-binding proteins. elav localization did not change notably upon VCP QQ expression (Fig. 4 E and F). Strikingly, TDP-43 became depleted from the cytoplasm of class IV da neurons and relocalized to the nucleus upon VCP QQ expression (Fig. 4 D and F). Closer inspection revealed that TDP-43 in VCP-inhibited neurons was now enriched in nuclear dots that often also exhibited increased elav staining. The relocalization of TDP-43 from the cytoplasm to the nucleus was also observed in vcp26-8 mutant class IV da neuron MARCM clones (Fig. 4 G–I), and with transgenic, HA-tagged TDP-43 upon VCP QQ expression (Fig. S1). Importantly, quantification and normalization of TDP-43 levels showed that VCP inhibition did not alter the absolute levels of TDP-43 (Fig. S1), suggesting that the observed effects were not a consequence of a defect in TDP-43 degradation. In fact, the only manipulation that resulted in a mild but significant increase in TDP-43 levels—but without a change in localization (Fig. S2)—was the expression of an RNAi directed against the autophagy factor ATG7, perhaps reflecting the degradation of cytoplasmic RNA granules through the autophagy pathway (27).
Fig. 4.
Links between VCP, RNA-binding proteins, and class IV da neuron dendrite pruning. (A–C) TDP-43 and elav localization in control class IV da neurons at the third instar larval stage. (A) TDP-43 is largely cytosolic in ddaC; (B) elav, a neuronal nuclear RNA-binding protein; and (C) merge. (D–F) TDP-43 and elav localization upon VCP QQ expression. (D) TDP-43 is now largely nuclear, (E) elav is not changed, and (F) the merge shows that TDP-43 now has a similar nuclear localization profile as elav. (G–I) TDP-43/elav localization in vcp26-8 mutant class IV da neurons: (G) TDP-43 staining, (H) elav staining, and (I) merge. vcp26-8 mutant class IV da neurons were generated by MARCM. In A–I, cell bodies of class IV da neurons are indicated by continuous lines and nuclei are marked by dashed lines. (J and K) Gain-of-function, but not loss-of function, of TDP-43 induces pruning defects at 16 h APF. (J) TDP-43 Q367X mutant class IV da neurons prune their dendrites normally (0/32 with dendrites still attached to the soma at 16 h APF), and (K) TDP-43 overexpression under ppk-GAL4 leads to pruning defects (23/24 with dendrites still attached to soma at 16 h APF, P < 0.005). (L) Synergistic effects of weak TDP-43 expression (UAS-HATDP-43weak) and VCP A229E. The indicated transgenes were expressed in class IV da neurons under the control of ppk-GAL4, and the percentage of neurons with unpruned dendrites was determined. Statistical analysis was by Fisher’s exact test (***P < 0.005). (M) RNAi-mediated elav down-regulation does not affect pruning (0/57 with attached dendrites). (N) elav overexpression inhibits pruning (51/55 with attached dendrites, P < 0.005). In J–M, class IV da neuron cell bodies are indicated by arrowheads. (Scale bars in A–I, 10 μm; in J–M, 20 μm.)
We next asked whether manipulation of TDP-43 would affect class IV da neuron dendrite pruning. A previously characterized TDP-43 mutant, TDP-43 Q367X (28), did not display pruning defects (Fig. 4J), but overexpression of TDP-43 (Fig. 4K) led to strong dendrite pruning defects at 16 h APF. In support of the hypothesis that TDP-43 acts in the same or a similar pathway as VCP during dendrite pruning, we also found that a more weakly expressed TDP-43 transgene (UAS–TDP-43weak) and VCP A229E, a weakly dominant-active VCP allele corresponding to a human VCP disease mutation (29), exhibited a synergistic inhibition of pruning when coexpressed (Fig. 4L). Interestingly, manipulation of elav gave very similar results as with TDP-43: elav down-regulation by RNAi did not affect pruning (Fig. 4M), but elav overexpression led to highly penetrant pruning defects (Fig. 4N).
To exclude the possibility that the pruning defects induced by TDP-43 or elav overexpression were due to long-term expression and aggregation of RNA-binding proteins, we also induced TDP-43 and elav overexpression acutely (24 h before the onset of pupariation). Pruning was still inhibited in these cases (Fig. S2). Also, overexpression of several other RNA-binding proteins did not cause pruning defects (Table S1), with two exceptions: a UAS-carrying P-element in the promotor of the adjacent x16 and HRP48 genes caused a strong pruning defect when expression was induced in class IV da neurons, and levels of a GFP protein trap insertion into the x16 gene were also markedly increased in class IV da neurons expressing VCP QQ (Fig. S2), possibly indicating a role for VCP in x16 degradation. In further support of an involvement of VCP with RNA-binding proteins during neuronal pruning processes, we also found that VCP is required for mushroom body γ neuron axon pruning and induces the accumulation of boule, an RNA-binding protein that had previously been shown to inhibit γ neuron axon pruning when overexpressed (30) (Fig. S3). Thus, our data suggest that VCP regulates a specific subset of RNA-binding proteins and that this regulatory role of VCP is associated with its role in pruning.
MICAL and TDP-43 Are Regulated by Ubiquitin and the 19S Proteasome.
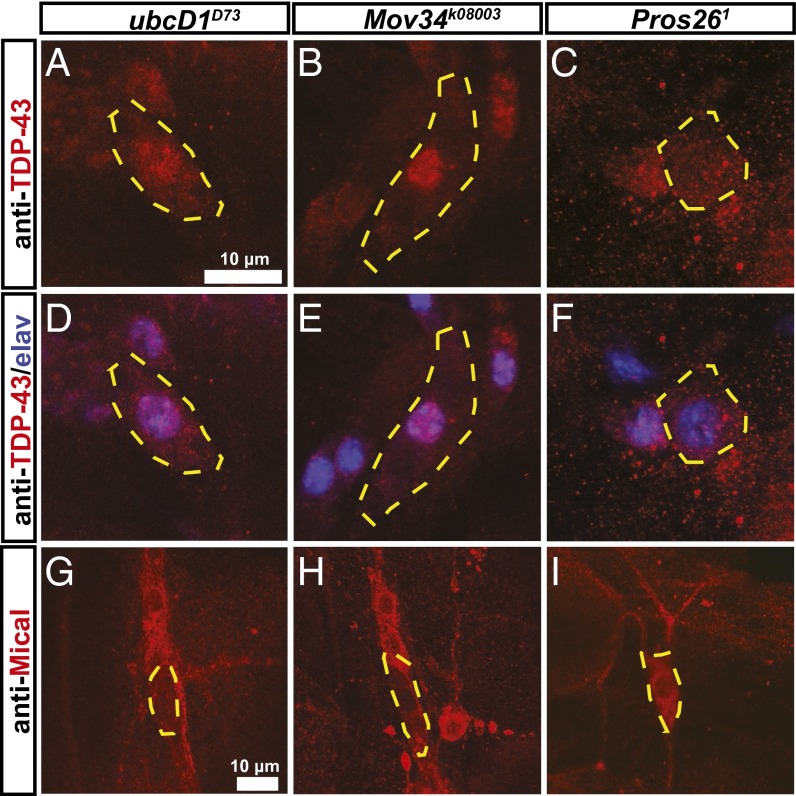
As VCP is an integral component of the UPS, we next asked whether the role of VCP in MICAL regulation and TDP-43 localization was also dependent on ubiquitylation and/or the proteasome. To address this question, we assessed Mical levels and TDP-43 distribution in UPS mutants with known pruning defects. An ubiquitylation enzyme known to be required for pruning is the E2 enzyme ubcD1 (7). When we assessed TDP-43 localization in larval ubcD1D73 mutant class IV da neurons, we found that TDP-43 was again localized to the nucleus in these cells (Fig. 5A). Furthermore, we noted a pronounced reduction of Mical expression in ubcD1D73 mutant class IV da neurons during the early pupal stage (Fig. 5D), indicating that ubiquitylation through ubcD1 is involved in the regulation of TDP-43 localization and Mical expression.
Fig. 5.
TDP-43 localization and Mical expression are regulated by ubiquitylation and the 19S proteasome. (A–F) TDP-43 and elav localization were assessed in MARCM clones of UPS mutant class IV da neurons at the third instar larval stage. Shown are TDP-43 localization (A–C) and merged TDP-43/elav stainings (D–F) in class IV da neuron MARCM clones mutant for ubcD1 (an E2 enzyme) (A and D), Mov34 (a 19S cap subunit) (B and E), and Pros26 (a 20S core subunit) (C and F). (A and D) TDP-43/elav localization in ubcD1D73 mutant ddaC neurons. (B and E) TDP-43/elav localization in Mov34k08003 mutant class IV da neurons. (C and F) TDP-43/elav localization in Pros261 mutant class IV da neurons. (G–I) Mical expression in class IV da neurons mutant for components of the UPS at 2 h APF: (G) ubcD1D73 mutant, (H) Mov34k08003 mutant, and (I) Pros261 mutant. Cell bodies of class IV da neurons are outlined.
We next assessed TDP-43 localization and Mical expression in proteasome mutants. We first used a previously characterized mutant in the Mov34 gene encoding the 19S subunit Rpn8 (8). TDP-43 was again relocalized to the nucleus in Mov34 mutant class IV da neurons (Fig. 5B), and Mical expression was absent from Mov34 mutant class IV da neurons at 2 h APF (Fig. 5H). To rigorously address whether proteasomal proteolysis was also required for TDP-43 localization and Mical expression, we next assessed the effect of Pros261, a previously characterized mutation in the 20S core particle subunit Prosβ6 (31). In contrast to Mov34 mutant class IV da neurons, Pros261 mutant class IV da neurons displayed cytoplasmic TDP-43 localization (Fig. 5C), and we detected robust Mical expression in these neurons at 2 h APF (Fig. 5I). Thus, although ubiquitylation and the 19S proteasome are both required for Mical expression and normal TDP-43 localization, proteolysis through the 20S core particle of the proteasome is not. Importantly, Pros261 MARCM class IV da neurons showed strong dendrite pruning defects at 16 h APF, as did expression of RNAi constructs directed against subunits of the 20S core particle (Fig. S4), thus showing that proteasomal proteolysis is required for dendrite pruning.
These data indicate that there must be several ubiquitin- and proteasome-dependent pathways that are required for dendrite pruning: one pathway requires ubcD1, VCP, and the 19S regulatory particle of the proteasome, but not the 20S core particle. This pathway regulates MICAL expression. A second UPS pruning pathway does depend on proteolysis through the 20S core. In an E3 ubiquitin ligase candidate screen, we identified cul-1/lin19 as a pruning mutant. Cul-1 encodes cullin-1, a core component of a class of multisubunit ubiquitin ligases known as SCF (for Skp1/Cullin/F-box) ligases (32). Class IV da neurons mutant for cul-1 or class IV da neurons expressing an RNAi construct directed against cul-1 had not pruned their dendrites at 16 h APF (Fig. S4). However, unlike with VCP, ubcD1, and Mov34, cul-1 mutation did not affect Mical expression at 2 h APF (Fig. S4), indicating that cullin-1 is not a component of the VCP-dependent UPS pathway involved in splicing and might thus be a component of a proteolytic UPS pathway. In support of this notion, a recent report independently identified cul-1 as a pruning mutant and associated it with protein degradation (33).
We had previously proposed that the E2 enzyme ubcD1 and VCP would act to activate caspases during pruning (7, 11). However, the dendrite pruning defects caused by those UPS mutants are much stronger than the phenotypes caused by caspase inactivation, which mostly causes a delay in the phagocytic uptake of severed dendrites by the epidermis (6, 34). Although we cannot exclude that ubcD1 and VCP contribute to caspase activation during pruning, the new mechanism proposed here—control of RNA-binding proteins and MICAL expression—likely makes a much stronger contribution to the drastic pruning phenotypes of UPS mutants.
How precisely do VCP, ubcD1, and the 19S proteasome contribute to MICAL expression? Our data indicate that VCP inhibition causes missplicing of MICAL mRNA that likely leads to the expression of an inactive Mical protein variant. At the same time, VCP inhibition leads to the mislocalization of TDP-43, and possibly the dysregulation of a number of other RNA-binding proteins. The fact that these phenotypes correlate in the vcp, ubcD1, and Mov34 mutants gives a strong indication that they are related. TDP-43 had previously been identified as a suppressor of the toxicity induced by a weak VCP disease allele in the Drosophila eye (21). In class IV da neurons, reducing the amounts of TDP-43 (with a deficiency) or elav (by RNAi) did not ameliorate the pruning defect induced by VCP inhibition [ppk > VCP QQ, 88% pruning defects (n = 35); ppk > VCP QQ, Df(2R)BSC660/+, 93% pruning defects (n = 16); ppk > VCP QQ, elavRNAi, 84% pruning defects (n = 12); P > 0.5 for each]. Therefore, we cannot exclude the possibility that the two proteins act in parallel rather than in an epistatic fashion. As VCP has been shown to remodel protein complexes that contain ubiquitylated proteins (12) and is structurally similar to the 19S cap, it is interesting to speculate that VCP and the 19S cap might alter the subunit composition of ubiquitylated TDP-43–containing complexes of RNA-binding proteins, and that this activity—rather than a direct action on TDP-43 (or maybe also elav) alone—might lead to both MICAL missplicing and TDP-43 mislocalization.
Interestingly, autosomal dominant mutations in human VCP cause frontotemporal dementia and ALS, a hallmark of which is the formation of aggregates that contain TDP-43. Most of these aggregates are cytoplasmic (and contain TDP-43 that has relocalized from the nucleus to the cytoplasm), but VCP mutations also induce TDP-43 aggregation in the nucleus (35), a situation that might be similar to the situation caused by VCP inhibition in class IV da neurons. Although human VCP disease mutations have been proposed to act as dominant-active versions of VCP with enhanced ATPase activity (29), both the disease allele and the dominant-negative ATPase-dead VCP QQ mutant cause class IV da neuron pruning defects and TDP-43 relocalization to the nucleus of class IV da neurons (Fig. S2) and therefore act in the same direction. It is thought that VCP can only bind substrates when bound to ATP, and will release bound substrates upon ATP hydrolysis (12). Thus, it is conceivable that the phenotypic outcome of inhibiting the ATPase (no substrate release) should be similar to that of ATPase overactivation (reduced substrate binding or premature substrate release): in both cases, a substrate protein complex would not be properly remodeled.
Taken together, our results indicate the existence of a nonproteolytic function of VCP and the UPS in RNA metabolism and highlight its importance during neuronal development.
Materials and Methods
Fly Stocks.
For labeling of class IV neurons, we used different ppk-GAL4 insertions on the second or third chromosome (36), ppk-GeneSwitch for inducible GAL4/UAS expression, or the enhancer fusion ppk-eGFP (37). GAL4NP21 was used to label mushroom body γ neurons. Constructs for RNA-binding proteins were UAS-TDP-43 (24), UAS-elav (Bloomington), and x16CB03248 (x16-GFP fusion, Flytrap); additional lines for RNA-binding proteins are listed in Table S1. Dominant-negative UAS-VCP QQ has been described (11). RNAi lines against elav, Prosα7, Prosβ5, and cul-1 were from Bloomington (TRiP JF03008, TRiP HMS00068, TRiP HMS00119, TRiP HM05197). RNAi experiments were done with coexpression of UAS-dcr2 (38). ATG7 IR was cloned into pWIZ, and Sox14 and HATDP-43 were cloned into pUAST attB (for ϕC31-mediated integration) and injected into flies carrying attP2, attP VK00037, or attP VK00016 acceptor sites. MARCM clones were generated by a modified procedure using SOP-FLP to induce mitotic recombination (39). Mutant chromosomes were FRT42D, vcp26-8/CyOweeP; FRT42D, P[lacW]Mov34k08003/CyOweeP (Bloomington) (3); FRT82B, ubcD1D73/Tm6b (8); FRT G13, P[lacW]lin19k01207/CyOweeP (8); FRT2A, Pros261/Tm6b; and TDP-43 Q367X (28).
Live Imaging.
Live imaging of da neurons in appropriately staged larvae or pupae was carried out on a Leica SP5 confocal microscope as described (11). Images were taken from neurons in abdominal segments A2–A6. In all images shown, anterior is left and dorsal is up. Fisher’s exact test (GraphPad) was used for statistical comparisons.
Antibodies and Immunocytochemistry.
Larvae or appropriately staged pupae were dissected as described (11). Antibodies used were rabbit–anti-Mical (1:3,000) (20), rabbit anti–dTDP-43 (1:200) (26), guinea pig anti-Sox14 (1:30) (40), mouse anti-Hdc (1:10) (41), rat anti-elav 7E8A10 (1:30) (Drosophila Studies Hybridoma Bank), rat anti-HA 3F10 (1:200), rat anti-mCD8 (1:200; Invitrogen), and chicken anti-GFP (1:200; Aves Laboratory). Cy2-, Cy5-, or Rhodamine Red X-conjugated secondary goat or donkey antibodies (Jackson Laboratory) were used at 1:100, 1:50, or 1:200, respectively.
FACS Analysis and RT-PCR.
Appropriately staged animals were dissected and digested with collagenase, and GFP-labeled ppk+ neurons were sorted with a 488 nm laser on a FACSAria II machine. RT-PCR was performed using the SuperScript III kit (Invitrogen) according to the manufacturer’s instructions using the primer pairs GCCAACTGCTTCTGATGGAGTCC and GAGCAGGAGAAGCTACATACC for exons 8–12 and GGACAAGCAGCTAAAGGAGGGC and GTTTGCTTACAGAAGCGGCAC for exons 14–16.
Supplementary Material
Acknowledgments
We are grateful to A. Kolodkin, C. K. Shen, F.-B. Gao, T. K. Sang, T. Uemura, A. Spradling, D. Rio, S. Wasserman, and N. Bonini for their kind gifts of fly lines and antibodies. We also thank the Bloomington Stock Center, the Vienna Drosophila RNAi Center, and the Drosophila Genomic Resource Center for fly lines and the Drosophila Studies Hybridoma Bank for antibodies. We thank S. Elmes and J. Gordon for help with FACS sorting, C. Tong for help with cloning, N. Ninov and M. Müller for critical reading of the manuscript, and Y.N.J. laboratory members for helpful comments. This work was supported by National Institutes of Health Grant R 37NS040929 (to Y.N.J.). L.Y.J. and Y.N.J. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406898111/-/DCSupplemental.
References
- 1.Luo L, O’Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- 2.Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA. 2005;102(42):15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: Insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132(16):3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 4.Kirilly D, et al. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci. 2009;12(12):1497–1505. doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA. 2009;106(15):6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9(10):1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 7.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51(3):283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: Evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38(6):871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 9.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 10.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rumpf S, Lee SB, Jan LY, Jan YN. Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development. 2011;138(6):1153–1160. doi: 10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jentsch S, Rumpf S. Cdc48 (p97): A “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32(1):6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JO, et al. ITALSGEN Consortium Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68(5):857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 16.Kanamori T, et al. Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science. 2013;340(6139):1475–1478. doi: 10.1126/science.1234879. [DOI] [PubMed] [Google Scholar]
- 17.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 18.Loncle N, Williams DW. An interaction screen identifies headcase as a regulator of large-scale pruning. J Neurosci. 2012;32(48):17086–17096. doi: 10.1523/JNEUROSCI.1391-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63(6):788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109(7):887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 21.Ritson GP, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30(22):7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou HL, Geng C, Luo G, Lou H. The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP. Genes Dev. 2013;27(9):1046–1058. doi: 10.1101/gad.215681.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 2001;15(19):2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin MJ, Cheng CW, Shen CK. Neuronal function and dysfunction of Drosophila dTDP. PLoS ONE. 2011;6(6):e20371. doi: 10.1371/journal.pone.0020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Ferris J, Gao FB. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol Brain. 2009;2:30–39. doi: 10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang YC, et al. Pathogenic VCP/TER94 alleles are dominant actives and contribute to neurodegeneration by altering cellular ATP level in a Drosophila IBMPFD model. PLoS Genet. 2011;7(2):e1001288. doi: 10.1371/journal.pgen.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoopfer ED, Penton A, Watts RJ, Luo L. Genomic analysis of Drosophila neuronal remodeling: A role for the RNA-binding protein Boule as a negative regulator of axon pruning. J Neurosci. 2008;28(24):6092–6103. doi: 10.1523/JNEUROSCI.0677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth KA, Belote JM. The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome beta-type subunit. Genetics. 1999;151(1):211–220. doi: 10.1093/genetics/151.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 33.Wong JJ, et al. A Cullin1-based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol. 2013;11(9):e1001657. doi: 10.1371/journal.pbio.1001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han C, et al. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron. 2014;81(3):544–560. doi: 10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann M. Molecular neuropathology of TDP-43 proteinopathies. Int J Mol Sci. 2009;10(1):232–246. doi: 10.3390/ijms10010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grueber WB, et al. Projections of Drosophila multidendritic neurons in the central nervous system: Links with peripheral dendrite morphology. Development. 2007;134(1):55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- 37.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13(8):618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 38.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 39.Matsubara D, Horiuchi SY, Shimono K, Usui T, Uemura T. The seven-pass transmembrane cadherin Flamingo controls dendritic self-avoidance via its binding to a LIM domain protein, Espinas, in Drosophila sensory neurons. Genes Dev. 2011;25(18):1982–1996. doi: 10.1101/gad.16531611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritter AR, Beckstead RB. Sox14 is required for transcriptional and developmental responses to 20-hydroxyecdysone at the onset of drosophila metamorphosis. Dev Dyn. 2010;239(10):2685–2694. doi: 10.1002/dvdy.22407. [DOI] [PubMed] [Google Scholar]
- 41.Weaver TA, White RA. headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development. 1995;121(12):4149–4160. doi: 10.1242/dev.121.12.4149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.