Significance
Circulating cell-free RNA in the blood provides a potential window into the health, phenotype, and developmental programs of a variety of human organs. We used high-throughput methods of RNA analysis such as microarrays and next-generation sequencing to characterize the global landscape of circulating RNA in human subjects. By focusing on tissue-specific genes, we were able to identify the relative contributions of these tissues to circulating RNA and monitor changes during tissue development and neurodegenerative disease states.
Keywords: noninvasive diagnostics, cell-free nucleic acids, genomics
Abstract
Circulating cell-free RNA in the blood provides a potential window into the health, phenotype, and developmental programs of a variety of human organs. We used high-throughput methods of RNA analysis such as microarrays and next-generation sequencing to characterize the global landscape circulating RNA in a cohort of human subjects. By focusing on genes whose expression is highly specific to certain tissues, we were able to identify the relative contributions of these tissues to circulating RNA and to monitor changes in tissue development and health. As one application of this approach, we performed a longitudinal study on pregnant women and analyzed their combined cell-free RNA transcriptomes across all three trimesters of pregnancy and after delivery. In addition to the analysis of mRNA, we observed and characterized noncoding species such as long noncoding RNA and circular RNA transcripts whose presence had not been previously observed in human plasma. We demonstrate that it is possible to track specific longitudinal phenotypic changes in both the mother and the fetus and that it is possible to directly measure transcripts from a variety of fetal tissues in the maternal blood sample. We also studied the role of neuron-specific transcripts in the blood of healthy adults and those suffering from the neurodegenerative disorder Alzheimer’s disease and showed that disease specific neural transcripts are present at increased levels in the blood of affected individuals. Characterization of the cell-free transcriptome in its entirety may thus provide broad insights into human health and development without the need for invasive tissue sampling.
The existence of cell-free nucleic acids—both DNA and RNA—in human blood has been known and studied since the late 1940s (1). These molecules are largely the debris of apoptotic and necrotic cells from different tissues that are released into blood and therefore represent a window into the health and status of many solid tissues. The utility of cell-free DNA has been powerfully demonstrated by the development of noninvasive prenatal tests for fetal aneuploidy (2), noninvasive sequencing of fetal genomes (3), noninvasive diagnostics for organ transplant recipients (4), and the ability to both diagnose cancer and monitor the response of cancer treatment (5–7). These approaches are rapidly moving beyond academic research and into clinical use. For example, it is estimated that in 2013 more than half a million pregnant women underwent noninvasive prenatal testing based on cell-free DNA technology (8). However, detecting differences in gene expression, and not just gene content, may improve our understanding and diagnosis of numerous disease states, including organ failure, inflammatory disease, neurodegenerative disorders, and additional pathological obstetric conditions.
In this respect, circulating cell-free RNA provides a unique opportunity to measure specific phenotypic information across many organs in the human body. Traditional molecular assays that accomplish this focus on either protein or metabolite measurements, which are challenging to perform on a large scale. Modern RNA analysis techniques such as microarrays and high-throughput sequencing provide the ability to make global measurements of RNA species and therefore enable one to probe a much more complex phenotypic landscape. Plasma cell-free RNA has been investigated in cancer patients (9–12) and maternal plasma (13, 14) and was assumed to be packaged in apoptotic bodies that give it stability in plasma (15). Studies of maternal plasma mRNA transcripts have focused mainly on transcripts that are exclusively expressed from the placenta (16–18) or on isolated plasma samples (19) and have not attempted to comprehensively study global fetal gene expression in vivo.
Results and Discussion
We used high-throughput methods of microarray and next-generation sequencing to characterize the landscape of cell-free RNA transcriptome of healthy adults and of pregnant women across all three trimesters of pregnancy and postpartum. Our results show that it is possible to monitor the gene expression status of many tissues and that one can measure the temporal expression of many crucial genes longitudinally during human development. In addition, our approach is able to quantitate various classes of transcripts, including long noncoding and circular RNA. Thus, cell-free RNA provides a unique in vivo window into the gene expression program of the developing human fetus. We also investigated the role of cell-free RNA in adults suffering from the neurodegenerative disorder Alzheimer’s syndrome and observed a marked increase of neuron-specific transcripts in the blood of affected individuals, thus suggesting that cell-free RNA may be a potential diagnostic tool for this disease.
In our initial study, plasma samples were collected from four nonpregnant subjects consisting of 2 men and 2 women, as well as from 11 pregnant women who contributed samples at the first, second, and third trimesters and postpartum (Fig. S1). After extraction from plasma, total mRNA was converted to cDNA and amplified, followed by sequencing. For each plasma sample, ∼20 million sequencing reads were generated, ∼80% of which could be mapped against the human reference genome (hg19). As the plasma RNA is of low concentration and vulnerable to degradation, contamination from the plasma DNA is a concern. To assess the quality of the sequencing library, the number of reads assigned to different regions was counted: 34% mapped to exons, 18% mapped to introns, and 24% mapped to ribosomal RNA and tRNA. Therefore, dominant portion of the reads originated from RNA transcripts rather than DNA contamination. To validate the RNA-seq measurements, all of the plasma samples were also analyzed with gene expression microarrays.
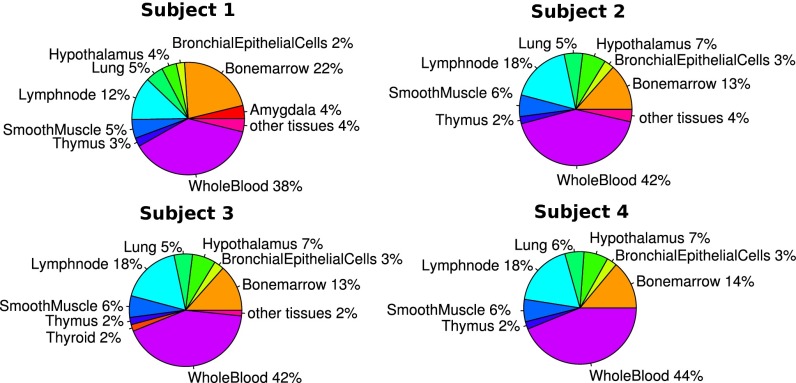
Apoptotic cells from different tissue types release their RNA into the cell-free RNA component in plasma. Each of these tissues expresses a number of genes unique to their tissue type, and the observed cell-free RNA transcriptomes can be considered as a summation of contributions from these different tissue types. Using expression data of different tissue types available in public databases, the cell-free RNA transcriptome from our four nonpregnant subjects were deconvoluted using quadratic programming to reveal the relative contributions of different tissue types (Fig. 1). These contributions identified different tissue types which are consistent among different control subjects. Whole blood, as expected, is the major contributor (∼40%) toward the cell-free RNA transcriptome. Other major contributing tissue types include the bone marrow and lymph nodes. One also sees consistent contributions from smooth muscle, epithelial cells, thymus, and hypothalamus.
Fig. 1.
Using microarray expression data from the plasma of four normal controls, quadratic programming was performed using known tissue-specific expression from a publicly available database to obtain the relative tissue contribution of the respective different tissue types. The relative contribution from detected tissues were represented in pie charts for these four subjects. Distribution of contribution from different tissue types are fairly consistent between these subjects, with the major contributor of cell-free RNA originating from whole blood.
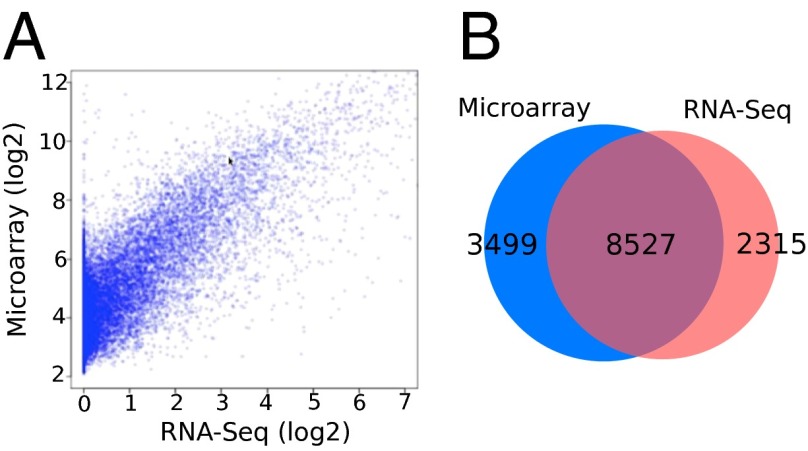
The transcriptome data obtained by both microarray and RNA-seq (20) were compared for each pregnant patient at various time points of gestation. Fig. 2 shows a scatter plot of the transcriptomes from one of the patients in the third trimester; both techniques showed good concordance with the Pearson correlation at 0.78. There are 8,527 detected genes (∼71% of all detected genes) that are common to both techniques (Table S1). An average Pearson correlation of 0.739 was obtained when considering all of the patients’ samples in our study (Table S2). About 15% of the transcripts detected by sequencing were long noncoding RNA, and the relative proportion of different ncRNA categories remained relatively stable and consistent across different patients and trimesters (Fig. S2).
Fig. 2.
Characterization of maternal plasma transcriptome by RNA-seq and microarray assays. (A) The scatter plot of the correlation between RNA-seq and Affymetrix array assay for samples taken at the third trimester. The Pearson correlation coefficient is 0.78. (B) Venn diagram displaying the genes detected by RNA-seq and microarray. The cutoff for the RNA-Seq is fragments per kilobase of transcript per million mapped reads (FPKM) > 0. The cutoff for the microarray is intensity > 4. Sample P12_T3 is shown here as an example.
Within our cohort, we analyzed ∼100 genes whose RNA transcripts contained paternal SNPs that were distinct from the maternal inheritance (Fig. S3) to explicitly demonstrate that the fetus contributes a substantial amount of RNA to the mother’s blood. To accurately quantify and verify the relative fetal contribution, we genotyped a mother and her fetus and inferred paternal genotype. We quantified the weighted average fraction of fetal-originated cell-free RNA using paternal SNPs. Cell-free RNA fetal fraction depends on gene expression and varies greatly across different genes. In general, the fetal fraction of cell-free RNA increases as the pregnancy progress and decreases after delivery. The weighted average fetal fraction started at 0.4% in the first trimester, increased to 3.4% in the second trimester, and peaked at 15.4% in the third trimester. (Figs. S4–S6) Although fetal RNA should be cleared after delivery, there was still 0.3% of fetal RNA as calculated, which can be attributed to background noise arising from misalignment and sequencing errors.
In addition to monitoring fetal tissue-specific mRNA, our approach also identifies noncoding transcripts present in the cell-free compartment across pregnancy. These noncoding transcripts include long noncoding RNAs (lncRNAs), as well as circular RNAs (21) (circRNA), which we show here to be detectable in the plasma (Table S3). Additional PCR assays were designed to specifically amplify and validate the presence of these circRNA in plasma (Fig. S7). circRNAs have recently been shown to be widely expressed in human cells (22) and have greater stability than their linear counterparts (23), potentially making them reliable biomarkers for capturing transient events. Several of the circRNA species appear to be specifically expressed during different trimesters of pregnancy (Fig. S8). The identification of these cell-free noncoding RNAs during pregnancy may improve our ability to monitor the health of the mother and fetus, especially as our understanding of the functions of these newly discovered RNA species matures.
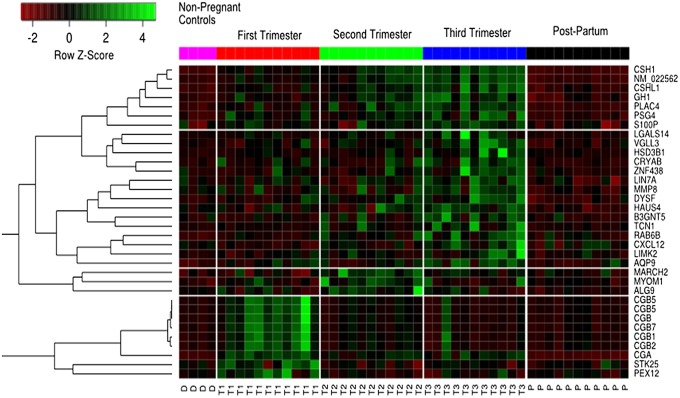
There is a general increase in the number of genes detected across the different trimesters followed by a steep drop after the pregnancy (Fig. S9). Such an increase in the number of genes detected suggests that we are observing unique transcripts expressed specifically during particular time intervals in the developing fetus. Fig. 3 and Fig. S10 show the heatmap of genes whose level changed over time during pregnancy, as detected by microarray and RNA-seq, respectively. ANOVA was applied to identify genes that varied in expression in a statistically significant manner across different trimesters. We imposed an additional condition filtering for transcripts that were expressed at low levels in both the postpartum plasma of pregnant subjects and in nonpregnant controls. Using these conditions, we identified 39 genes from RNA-seq and 34 genes from microarray, of which there were 17 genes in common. Gene Ontology (GO) performed on the identified genes using Database for Annotation, Visualization and Integrated Discovery (DAVID) (24) revealed that the identified gene list is enriched for the following GO terms: female pregnancy (Bonferroni-corrected P = 5.5 × 10−5), extracellular region (corrected P = 6.6 × 10−3), and hormone activity (corrected P = 6.3 × 10−9). We also observed that these RNA transcripts show a general trend of having low expression postpartum and the highest expression during the third trimester. Most of these transcripts are specifically expressed in the placenta, and their levels reach a maximum in the later stages of pregnancy.
Fig. 3.
Heatmap of time-varying genes identified from microarray analysis. The color bar on the top of the heatmap corresponds to different time points during pregnancy. Each row of the heatmap refers to a gene, and each column is a sample taken at a particular time point: D, donor; T1, first trimester; T2, second trimester; T3, third trimester; P, postpartum. Unsupervised clustering was performed on genes across different time points. Distinct temporal trends are observed; the cluster of genes belongs to the CGB family of genes, which are known to be expressed at high levels during the first trimester exhibited corresponding high levels of RNA in the first trimester. The other clusters of genes mainly contains genes transcripts that increased throughout the first two trimesters and showed a peak expression in the third.
One of the most distinct temporal trends is that exhibited by the chorionic gonadotropin (CG) family of RNA transcripts. This family of transcripts is produced by placental syncytiotrophoblasts, and the maternal serum level of beta human chorionic gonadotropin (bHCG) has been previously shown to peak during the first trimester of pregnancy (25). This trend is similarly reflected within the cell-free transcriptome. Detection of bHCG protein in the blood and urine is routinely used to diagnose pregnancies and to monitor germ cell tumors and gestational trophoblastic disease. The fact that the RNA transcript level shows the same trends as the protein supports the notion of using RNA as a practical biomarker in situations where it is not straightforward to detect the protein (26). Along with HCG, other placental transcripts such as PLAC4, PSG4, HAUS4, GH1, and CSHL1 were also identified. These transcripts all share a similar temporal trend where we observed an increasing trend during pregnancy with the peak achieved during third trimester of pregnancy.
We identified other nonplacental transcripts that share similar temporal trends. Two such significant transcripts were RAB6B and MARCH2, which are known to be expressed specifically in CD71+ erythrocytes. Erythrocytes enriched for CD71+ have been shown to contain fetal hemoglobin and are interpreted to be of fetal origin (27). The presence of transcripts with known specificity to different fetal tissue types reflects the fact that the cell-free transcriptome during the period of pregnancy can be considered as a summation of transcriptomes from various different fetal tissues on top of a maternal background.
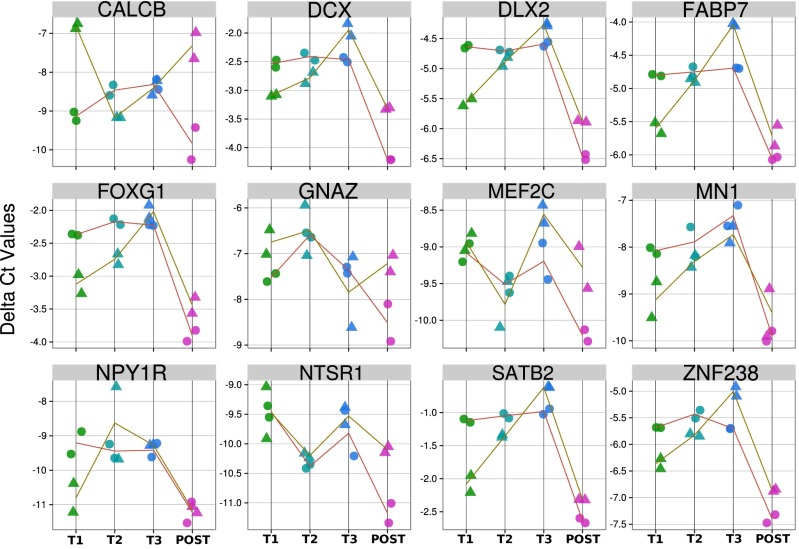
This analysis detected the presence of numerous transcripts that are specifically expressed in several other fetal tissues, although the available sequencing depth resulted in limited concordance between samples. To verify the presence of these and other potential fetal tissue-specific transcripts, we devised a panel of fetal tissue-specific transcripts for detailed quantification using the more sensitive method of quantitative PCR (qPCR). We focused on three main sources, which are of interest to fetal neurodevelopment and metabolism: placenta, fetal brain, and fetal liver. In Fig. 4 and Figs. S11 and S12, we systematically compare the levels of these groups of fetal tissue-specific transcripts at different trimesters to the level seen in maternal serum after delivery. To illustrate the temporal trends, we used housekeeping genes as the baseline, and ΔCt analysis was applied to find the level of relative expression these fetal tissue-specific transcripts with respect to the housekeeping genes. We observed that many of these tissue-specific transcripts were expressed at substantially higher levels during the pregnancy compared with postpartum. We found a general trend of an increase in the quantity of these transcripts across advancing gestation.
Fig. 4.
Time course of fetal brain-specific genes, measured by qPCR. Plot showing the ΔCt value with respect to the housekeeping gene ACTB across the different trimesters of pregnancy including after birth. Time points across each patient is shown connected by the lines. Two replicates were performed for each patient at each time point. The general trends show elevated levels during the trimesters with a decline to low levels after the baby is born in concordance with the notion that fetal-specific transcripts increased into the pregnancy followed by rapid clearance after birth.
The placental qPCR assay focused on genes that are known to be highly expressed in the placenta, many of which encode for proteins that have been shown to be present in the maternal blood. The serum levels of these proteins are known to be involved in pregnancy complications such as preeclampsia and premature births (28). Examples in our panel includes ADAM12, which encodes for disintegrin, and metalloproteinase domain-containing protein 12. These proteinases are highly expressed in human placenta and are present at high concentrations in maternal serum as early as the first trimester (29). ADAM12 serum concentrations are known to be significantly reduced in pregnancies complicated by fetal trisomy 18 and trisomy 21 and may therefore be of potential use in conjunction with cell-free DNA for the detection of chromosomal abnormalities (30, 31). Similarly, placental alkaline phosphatase, encoded by the ALPP gene, is a tissue-specific isoform expressed increasingly throughout pregnancy until term in the placenta (32, 33). It is anchored to the plasma membrane of the syncytiotrophoblast and to a lesser extent of cytotrophoblastic cells (34). This enzyme is also released into maternal serum (35), and variations of its concentration are related with several clinical disorders such as preterm delivery (36, 37). Another gene in our panel, BACE2, encoded the β site APP-cleaving enzyme, which generates amyloid-β protein by endoproteolytic processing. Brain deposition of amyloid-β protein is a frequent complication of Down syndrome patients, and BACE-2 is known to be overexpressed in Down syndrome (38).
Other transcripts in our placental assay are known to be transcribed at high levels in the placenta, and levels of these mRNAs are important for normal placental function and development in pregnancy. TAC3 is mainly expressed in the placenta and is significantly elevated in preeclamptic human placentas at term (39, 40). Similarly, PLAC1 is essential for normal placental development. PLAC1 deficiency results in a hyperplastic placenta, characterized by an enlarged and dysmorphic junctional zone (41). An increase in cell-free mRNA of PLAC1 has been suggested to be correlated with the occurrence of preeclampsia (42).
On the fetal liver tissue-specific panel, one of the characterized transcripts is AFP. AFP encodes for α-fetoprotein and is transcribed mainly in the fetal liver. AFP is the most abundant plasma protein found in the human fetus (43). Clinically, AFP protein levels are measured in pregnant women in either maternal blood or amniotic fluid and serve as a screening marker for fetal aneuploidy, as well as neural tube and abdominal wall defects (44, 45). Other fetal liver-specific transcripts that were characterized are highly involved in metabolism. An example is fetal liver-specific monooxygenase CYP3A7, which catalyzes many reactions involved in synthesis of cholesterol and steroids and is responsible for the metabolism of more than 50% of all clinical pharmaceuticals. In drug-treated diabetic pregnancies in which glucose levels in the woman are uncontrolled, neural tube and cardiac defects in the early developing brain, spine, and heart depend on functional GLUT2 carriers, whose transcripts are well characterized in our panel (46, 47). Mutations in this gene results in Fanconi–Bickel syndrome, a congenital defect of facilitative glucose transport (48). Monitoring of fetal liver-specific transcripts during the drug regime may enable analysis of the fetuses' response to drug therapy that the mother is undergoing.
The fetal brain panel focused on genes whose transcripts are brain specific and known to exhibit an increasing trend throughout development; many are also found within the amniotic fluid (49). An example is ZNF238, which is specific to fetal brain tissue and is known to be vital for cerebral cortex expansion during embryogenesis when neuronal layers are formed. Loss of ZNF238 in the central nervous system leads to severe disruption of neurogenesis resulting in a striking postnatal small-brain phenotype (50). Similarly, FOXG1 encodes a brain-specific transcriptional repressor that is essential for early development of the telencephalon (51) and is implicated in the severe neurodevelopmental disease Rett syndrome (52). The ability to measure expression of transcription factors enables one to directly monitor the controlling elements of the developmental program of human fetus in vivo and noninvasively.
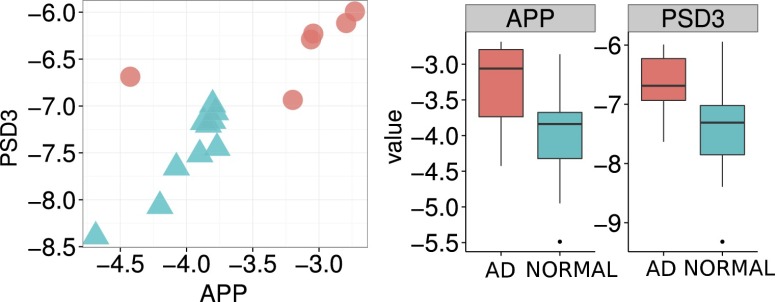
It was striking that we could detect brain-specific transcripts in multiple ways. The qPCR brain panel detected fetal brain-specific transcripts in maternal blood, whereas the whole transcriptome deconvolution analysis in our nonpregnant adult samples revealed the hypothalamus as a significant contributor. However, the fact that the hypothalamus is bounded by specialized brain regions that lack an effective blood–brain barrier (53) rationalizes these observations and leads one to hypothesize that cell-free DNA in the blood can be used to measure neuronal death. To follow up on this idea, qPCR was used to measure the expression levels of selected brain transcripts in the plasma of both Alzheimer's patients and age-matched normal controls. These measurements were made for a cohort of 16 patients: 6 diagnosed as Alzheimer's and 10 normal subjects (Fig. 5). The APP transcript encodes for the precursor molecule whose proteolysis generates β amyloid, which is the primary component of amyloid plaques found in the brain of Alzheimer's disease patients (54). Preliminary measurements of the plasma APP transcript corroborate the known biology behind progression of Alzheimer's disease (55–58) and showed a significant increase in patients with Alzheimer’s disease compared with normal subjects, suggesting that plasma APP mRNA levels may be a good marker for diagnosing Alzheimer's disease. Similarly, the gene PSD3, which is highly expressed in the nervous system and localized to the postsynaptic density based on sequence similarities (59, 60), shows an increase in the plasma of patients with Alzheimer's disease. By plotting the ΔCt values of APP against PSD3, we observed clustering of AD patients away from the normal patients. Although the ultimate sensitivity and specificity of this approach remain to be determined, it is intriguing to consider that cell-free RNA may serve as a potential blood test-based diagnostic for Alzheimer’s disease and other neurodegenerative disorders.
Fig. 5.
Measurements of PSD3 and APP cell-free RNA transcripts levels in plasma shows that the levels of these two transcripts are elevated in AD patients and can be used to cleanly group the AD patients from the normal patients.
Regulation of gene expression in space and time is key to understanding how the underlying complexity and variation during the process of fetal development can arise from the DNA blueprint. Until now, few studies have been able to capture the temporal dynamics of gene transcription in human fetuses during the course of pregnancy. Here, we discovered plasma cell-free transcripts that exhibit temporal dynamics during pregnancy. We characterized the dynamics underlying the maternal transcriptome and extracted fetal tissue-specific information. We anticipate these results are a stepping stone toward translating the temporal dynamics of plasma mRNA for clinical diagnosis of pregnancy-associated complications and developmental diseases, especially those that are temporal in nature and involve cellular apoptosis.
Materials and Methods
Detailed information about the analysis procedures of the experimental data for qRT-PCR, sequencing, and microarray can be found in SI Materials and Methods.
Clinical Sample Collection.
Following informed consent, pregnant patients were recruited in a Stanford University Institutional Review Board-approved protocol. Peripheral blood was prospectively obtained during each trimester during the course of pregnancy and within 6 wk postpartum. Samples for nonpregnant control patients were obtained from a Stanford University Institutional Review Board-approved blood bank protocol. For the cell-free RNA measurements of patients with Alzheimer’s disease, plasma samples were drawn from six clinically diagnosed patients with Alzheimer’s disease and an additional 10 age-matched controls.
Extraction of Cell-Free RNA from Plasma.
Patient blood was collected into EDTA-coated Vacutainers. Blood was centrifuged at 1,600 × g for 10 min at 4 °C, and the plasma was centrifuged again at 16,000 × g for 10 min at 4 °C to remove residual cells. Cell-free RNA was extracted from 3 mL of plasma using TRIzol (Invitrogen) and the RNeasy Kit (Qiagen) with DNase I digestion according to the manufacturer’s instructions. RNA integrity was verified using the RNA 6000 Pico Kit on an Agilent Bioanalyzer 2100 (Agilent).
cDNA Synthesis and Amplification.
cDNA was synthesized and amplified from cell-free RNA using the NuGen’s RNA-Seq Ovation System Kit (NuGen). The amplified cDNA was quantified by real-time PCR using TaqMan Universal Master Mix (ABI) on a Stratagen Real-Time PCR System (Stratagene). The probes of the real-time PCR were targeting to one housekeeping gene, GAPDH, and one pregnancy-specific gene, PLAC4.
Deconvolution of Cell-Free RNA Transcriptome Using Microarray.
Deconvolution of a cell-free transcriptome is used to determine the relative contribution of each tissue type toward the cell-free RNA transcriptome. The following steps are used to determine the relative RNA contributions of certain tissues in a sample. First, tissue-specific gene expression is identified using the Human 133A/GNF1H Gene Atlas Database. Microarray measurements of the cell-free RNA in plasma from a sample is assessed against these known tissue-specific expression profiles. The total RNA is considered a summation of these different tissue-specific transcripts. Quadratic programming can be used as a constrained optimization method to deduce the relative optimal contributions of different organs/tissues toward the cell-free transcriptome of the sample
where Y is the observed cell-free transcriptome; π is the fractional contribution of the tissue type toward the observed maternal trancriptome; X is the known tissue-specific gene expression vector for that particular tissue; ε is the normally distributed error; and i is the index of different tissue type. Additional constraints includes ∑πi = 1 and πi ≥ 0, because π is defined as the fractional contribution of each tissue type. Consequently, to obtain the optimal fractional contribution of each tissue type, we seek to minimize least-square error subjected to stated constraints. The above equations are then implemented using quadratic programming in R to obtain the relative contributions of the tissue types toward the cell-free transcriptome.
Parallel qPCR for Panel of Selected Transcripts.
Quantification of these fetal tissue-specific transcripts was carried out using the EvaGreen (Biotium) chemistry on the Fluidigm Biomark system (61). This system allows for simultaneous query of a panel of fetal tissue-specific transcripts. Amplification was performed using Evagreen primers targeting the genes of interest. Extracted RNA source was preamplified using the CellsDirect One-Step qRT-PCR kit (Invitrogen). Modifications were made to the default One-Step qRT-PCR protocol to accommodate a longer incubation time for reverse transcription. Nineteen cycles of preamplification were conducted, and the collected PCR products were cleaned up using exonuclease I treatment. To increase the dynamic range and the ability to quantify the efficiency of the later qPCR steps, serial dilutions were performed on the PCR products across 5-, 10-, and 20-fold dilutions. Each of the collected maternal plasma samples from individual pregnant woman across the time points went through the same procedures and was loaded onto 48 × 48 dynamic array chips (Fluidigm) to perform the qPCR. For a positive control, fetal tissue-specific RNAs from the various fetal tissue types were purchased from Agilent. Each of these RNAs from fetal tissues went through the same preamplification and exonuclease treatment. All collected data from the Fluidigm system were preprocessed using Fluidigm real-time PCR analysis software to obtain the respective Ct values for each of the transcripts across all samples. Negative controls of the experiments were performed for the entire process with water, as well as with samples that had not undergone the reverse transcription process.
Analysis of Ct Values.
Prior studies have shown that fetal tissue-specific RNA transcripts will be cleared from the maternal peripheral bloodstream within a short period after birth (16). Hence, the cell-free RNA repertoire after birth will be lacking in fetal tissue-specific RNA transcripts. We can then expect that the quantity of these fetal tissue-specific transcripts to be higher than that after birth. We are interested in the relative quantitative changes of the tissue-specific transcripts across all three trimesters of pregnancy compared with the baseline level after the baby is born. Analysis was conducted by comparing the fold change levels of each of these fetal tissue-specific transcripts across all three trimesters using the housekeeping genes as the baseline for comparison; we verified from the microarray and RNAseq data that the levels of the housekeeping genes did not significantly change throughout the course of pregnancy or postpartum. The ΔCt method is used here (62). Subsequently, the ΔCt value can be interpreted as a measure of the level of expression of the transcripts across each trimesters to postpartum and is plotted against the trimesters to illustrate the temporal trends.
Detection of Circular RNA Transcripts.
Detection was performed using the computation pipeline described in Memczak et al. on the sequenced data of maternal plasma (22).
Supplementary Material
Acknowledgments
We thank Norma Neff for assistance with sequencing and Lance Martin for helpful discussions. This work was supported in part by the March of Dimes. W.K. was supported by a fellowship from the Agency for Science, Technology and Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: The gene expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo(accession no. GSE56899).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405528111/-/DCSupplemental.
References
- 1.Mandel P, Métais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan HC, et al. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487(7407):320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci USA. 2011;108(15):6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100(15):8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leary RJ, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi DW, et al. In case you missed it: The Prenatal Diagnosis section editors bring you the most significant advances of 2013. Prenat Diagn. 2014;34(1):1–5. doi: 10.1002/pd.4288. [DOI] [PubMed] [Google Scholar]
- 9.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5(8):1961–1965. [PubMed] [Google Scholar]
- 10.Lo K-W, et al. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45(8 Pt 1):1292–1294. [PubMed] [Google Scholar]
- 11.Chen XQ, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6(10):3823–3826. [PubMed] [Google Scholar]
- 12.García-Olmo D, García-Olmo DC, Ontañón J, Martinez E, Vallejo M. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol. 1999;14(4):1159–1164. doi: 10.14670/HH-14.1159. [DOI] [PubMed] [Google Scholar]
- 13.Poon LLM, Leung TN, Lau TK, Lo YMD. Presence of fetal RNA in maternal plasma. Clin Chem. 2000;46(11):1832–1834. [PubMed] [Google Scholar]
- 14.Tsui NBY, Ng EKO, Lo YMD. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48(10):1647–1653. [PubMed] [Google Scholar]
- 15.Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47(8):1488–1489. [PubMed] [Google Scholar]
- 16.Ng EKO, et al. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100(8):4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo YMD, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat Med. 2007;13(2):218–223. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 18.Ng EKO, et al. The concentration of circulating corticotropin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin Chem. 2003;49(5):727–731. doi: 10.1373/49.5.727. [DOI] [PubMed] [Google Scholar]
- 19.Maron JL, et al. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J Clin Invest. 2007;117(10):3007–3019. doi: 10.1172/JCI29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 23.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Licht P, Russu V, Wildt L. On the role of human chorionic gonadotropin (hCG) in the embryo-endometrial microenvironment: Implications for differentiation and implantation. Semin Reprod Med. 2001;19(1):37–47. doi: 10.1055/s-2001-13909. [DOI] [PubMed] [Google Scholar]
- 26.Krege S, et al. European consensus conference on diagnosis and treatment of germ cell cancer: A report of the second meeting of the European Germ Cell Cancer Consensus group (EGCCCG): Part I. Eur Urol. 2008;53(3):478–496. doi: 10.1016/j.eururo.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Høgh AM, et al. zeta-, epsilon-, and gamma-Globin mRNA in blood samples and CD71(+) cell fractions from fetuses and from pregnant and nonpregnant women, with special attention to identification of fetal erythroblasts. Clin Chem. 2001;47(4):645–653. [PubMed] [Google Scholar]
- 28.Than NG, et al. Placental protein 13 (galectin-13) has decreased placental expression but increased shedding and maternal serum concentrations in patients presenting with preterm pre-eclampsia and HELLP syndrome. Virchows Arch. 2008;453(4):387–400. doi: 10.1007/s00428-008-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40(9):1685–1702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Laigaard J, et al. The level of ADAM12-S in maternal serum is an early first-trimester marker of fetal trisomy 18. Prenat Diagn. 2005;25(1):45–46. doi: 10.1002/pd.1029. [DOI] [PubMed] [Google Scholar]
- 31.Ito N, et al. ADAMs, a disintegrin and metalloproteinases, mediate shedding of oxytocinase. Biochem Biophys Res Commun. 2004;314(4):1008–1013. doi: 10.1016/j.bbrc.2003.12.183. [DOI] [PubMed] [Google Scholar]
- 32.Fishman L, Miyayama H, Driscoll SG, Fishman WH. Developmental phase-specific alkaline phosphatase isoenzymes of human placenta and their occurrence in human cancer. Cancer Res. 1976;36(7 PT 1):2268–2273. [PubMed] [Google Scholar]
- 33.Okamoto T, et al. Expression of human placenta alkaline phosphatase in placenta during pregnancy. Placenta. 1990;11(4):319–327. doi: 10.1016/s0143-4004(05)80223-1. [DOI] [PubMed] [Google Scholar]
- 34.Leitner K, et al. Placental alkaline phosphatase expression at the apical and basal plasma membrane in term villous trophoblasts. J Histochem Cytochem. 2001;49(9):1155–1164. doi: 10.1177/002215540104900909. [DOI] [PubMed] [Google Scholar]
- 35.Fishman WH, Ghosh NK, Inglis NR, Green S. Quantitation of the placental isoenzyme of alkaline phosphatase in pregnancy sera. Enzymologia. 1968;34(5):317–321. [PubMed] [Google Scholar]
- 36.Meyer RE, Thompson SJ, Addy CL, Garrison CZ, Best RG. Maternal serum placental alkaline phosphatase level and risk for preterm delivery. Am J Obstet Gynecol. 1995;173(1):181–186. doi: 10.1016/0002-9378(95)90187-6. [DOI] [PubMed] [Google Scholar]
- 37.Moawad AH, et al. NICHD MFMU Network The Preterm Prediction Study: The value of serum alkaline phosphatase, alpha-fetoprotein, plasma corticotropin-releasing hormone, and other serum markers for the prediction of spontaneous preterm birth. Am J Obstet Gynecol. 2002;186(5):990–996. doi: 10.1067/mob.2002.121727. [DOI] [PubMed] [Google Scholar]
- 38.Barbiero L, et al. BACE-2 is overexpressed in Down’s syndrome. Exp Neurol. 2003;182(2):335–345. doi: 10.1016/s0014-4886(03)00049-9. [DOI] [PubMed] [Google Scholar]
- 39.Page NM, Dakour J, Morrish DW. Gene regulation of neurokinin B and its receptor NK3 in late pregnancy and pre-eclampsia. Mol Hum Reprod. 2006;12(7):427–433. doi: 10.1093/molehr/gal025. [DOI] [PubMed] [Google Scholar]
- 40.Schlembach D, et al. Neurokinin B peptide serum levels are higher in normotensive pregnant women than in preeclamptic pregnant women. Am J Obstet Gynecol. 2003;189(5):1418–1422. doi: 10.1067/s0002-9378(03)00775-0. [DOI] [PubMed] [Google Scholar]
- 41.Jackman SM, Kong X, Fant ME. Plac1 (placenta-specific 1) is essential for normal placental and embryonic development. Mol Reprod Dev. 2012;79(8):564–572. doi: 10.1002/mrd.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purwosunu Y, et al. Cell-free mRNA concentrations of CRH, PLAC1, and selectin-P are increased in the plasma of pregnant women with preeclampsia. Prenat Diagn. 2007;27(8):772–777. doi: 10.1002/pd.1780. [DOI] [PubMed] [Google Scholar]
- 43.Ruoslahti E, Pihko H, Seppälä M. Alpha-fetoprotein: Immunochemical purification and chemical properties. Expression in normal state and in malignant and non-malignant liver disease. Transplant Rev. 1974;20(0):38–60. doi: 10.1111/j.1600-065x.1974.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 44.Rosen T, D’Alton ME. Down syndrome screening in the first and second trimesters: What do the data show? Semin Perinatol. 2005;29(6):367–375. doi: 10.1053/j.semperi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Bredaki FE, Poon LC, Birdir C, Escalante D, Nicolaides KH. First-trimester screening for neural tube defects using alpha-fetoprotein. Fetal Diagn Ther. 2012;31(2):109–114. doi: 10.1159/000335677. [DOI] [PubMed] [Google Scholar]
- 46.Li R, Thorens B, Loeken MR. Expression of the gene encoding the high-Km glucose transporter 2 by the early postimplantation mouse embryo is essential for neural tube defects associated with diabetic embryopathy. Diabetologia. 2007;50(3):682–689. doi: 10.1007/s00125-006-0579-7. [DOI] [PubMed] [Google Scholar]
- 47.Moley KH, Chi MM, Knudson CM, Korsmeyer SJ, Mueckler MM. Hyperglycemia induces apoptosis in pre-implantation embryos through cell death effector pathways. Nat Med. 1998;4(12):1421–1424. doi: 10.1038/4013. [DOI] [PubMed] [Google Scholar]
- 48.Santer R, et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17(3):324–326. doi: 10.1038/ng1197-324. [DOI] [PubMed] [Google Scholar]
- 49.Hui L, Slonim DK, Wick HC, Johnson KL, Bianchi DW. The amniotic fluid transcriptome: A source of novel information about human fetal development. Obstet Gynecol. 2012;119(1):111–118. doi: 10.1097/AOG.0b013e31823d4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang C, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19(4):692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303(5654):56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 52.Ariani F, et al. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008;83(1):89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fry M, Ferguson AV. The sensory circumventricular organs: Brain targets for circulating signals controlling ingestive behavior. Physiol Behav. 2007;91(4):413–423. doi: 10.1016/j.physbeh.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Matsui T, et al. Expression of APP pathway mRNAs and proteins in Alzheimer’s disease. Brain Res. 2007;1161:116–123. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 55.Su JH, Anderson AJ, Cummings BJ, Cotman CW. Immunohistochemical evidence for apoptosis in Alzheimer’s disease. Neuroreport. 1994;5(18):2529–2533. doi: 10.1097/00001756-199412000-00031. [DOI] [PubMed] [Google Scholar]
- 56.Estus S, et al. Aggregated amyloid-beta protein induces cortical neuronal apoptosis and concomitant “apoptotic” pattern of gene induction. J Neurosci. 1997;17(20):7736–7745. doi: 10.1523/JNEUROSCI.17-20-07736.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imaizumi K, et al. The cell death-promoting gene DP5, which interacts with the BCL2 family, is induced during neuronal apoptosis following exposure to amyloid beta protein. J Biol Chem. 1999;274(12):7975–7981. doi: 10.1074/jbc.274.12.7975. [DOI] [PubMed] [Google Scholar]
- 58.Mattson MP. Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol. 2000;1(2):120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 59.Nagase T, et al. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6(1):63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- 60.Nakajima D, et al. Construction of expression-ready cDNA clones for KIAA genes: Manual curation of 330 KIAA cDNA clones. DNA Res. 2002;9(3):99–106. doi: 10.1093/dnares/9.3.99. [DOI] [PubMed] [Google Scholar]
- 61.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE. 2008;3(2):e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.