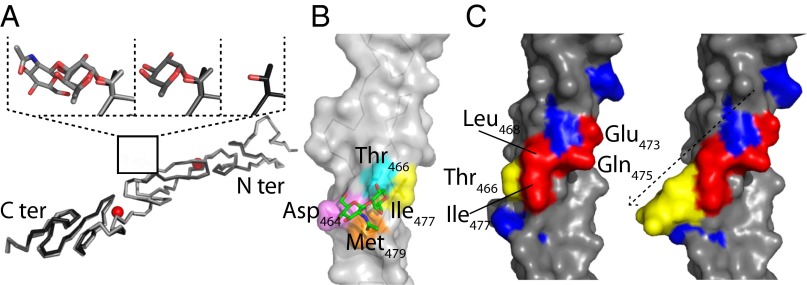
Fig. 4.
Structure of O-fucosylated variants of hN111–13. (A) Superimposed X-ray structures of the unmodified hN111–13 (PDB ID code 2VJ3) and the monosaccharide and disaccharide structures with the subsequent additions to the T466 region highlighted. Ca2+ ions are shown in red. (B) hN112 disaccharide X-ray structure highlighting contacts between the C6 methyl group of the O-fucose with Ile477 (yellow) and Met479 (orange), the C6 methoxy group of GlcNAc with Asp464 (pink), and the N-acetyl group of GlcNAc with Met479 (orange). Thr466 is highlighted in cyan. (C) hN112 unmodified and disaccharide X-ray structures highlighting residues shown to contribute to Jagged1 binding in a previous mutagenesis study (red); those residues that did not contribute to binding are also shown (blue) (32). T466 is highlighted in yellow, together with the disaccharide; the sugar may extend the known ligand-binding surface or act indirectly to promote complex formation.