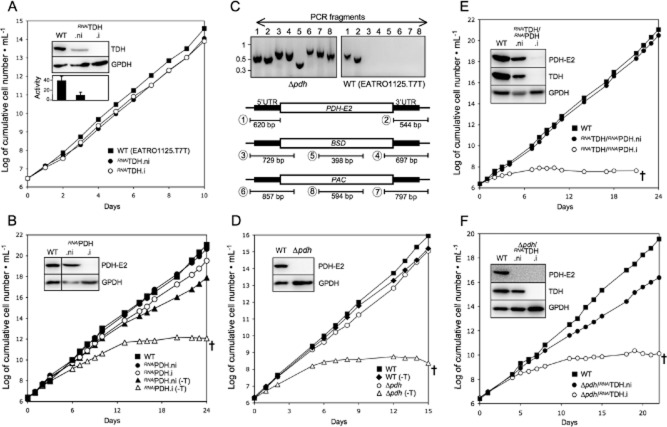
Figure 5.
Analysis of mutant cell lines. The figure shows growth curve of the Δpdh null mutant (D) and of the RNAiTDH (A), RNAiPDH (B), RNAiTDH/RNAiPDH (E) and Δpdh/RNAiTDH (F) mutant cell lines incubated in the presence (.i, ○) or in the absence (.ni, •) of tetracycline, and compared with the parental EATRO1125.T7T cell line (WT, ▪). Cells were maintained in the exponential growth phase (between 106 and 107 cells ml−1) and cumulative cell numbers reflect normalization for dilution during cultivation. All cell lines were grown in SDM79 medium; the RNAiPDH (B), Δpdh (D) and WT (D) cell lines were also grown in threonine-depleted conditions (-T). Crosses indicate that all cells were dead. The insets in panels A–B and D–F show Western blot analyses of the parental (WT) and mutant cell lines with the immune sera indicated in the right margin. The lower inset in panel A shows the TDH activity (milliunits mg−1 of protein) normalized with the malic enzyme activity measured in the same samples. Panel C, shows a PCR analysis of genomic DNA isolated from the parental EATRO1125.T7T (WT) and Δpdh cell lines. Amplifications were performed with primers based on sequences that flank the 5′UTR and 3′UTR fragments used to target the PDH-E2 gene depletion (black boxes) and internal sequences from the PDH-E2 gene (PCR products 1 and 2), the blasticidin resistance gene (BSD, PCR products 3–5) or the puromycin resistance gene (PAC, PCR products 6–8). As expected, PCR amplification using primers derived from the PDH-E2 gene and drug resistant genes were only observed for the parental EATRO1125.T7T and Δpdh cell lines respectively.