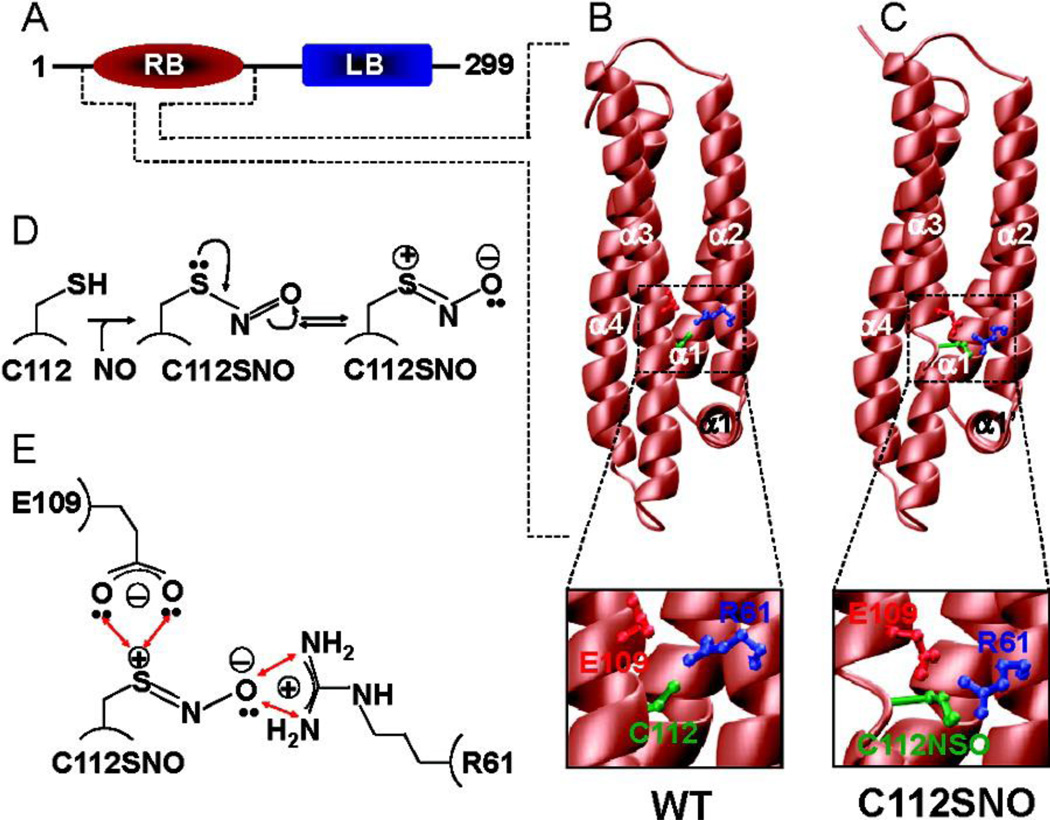
Figure 2. Effect of S-nitrosylation on the 3D structure of human ApoE3.
(A) Fully processed ApoE3, without the N-terminal signal peptide sequence (18 residues), is comprised of an N-terminal LDL receptor binding (RB) domain and a C-terminal lipid binding (LB) domain. Note that all the amino acid numbering used here is based on the amino acid sequence of the fully processed ApoE (residues 1−299). (B) 3D atomic model of the WT RB domain of ApoE. (C) 3D atomic model of the S-nitrosothiol derivative (C112SNO) of the RB domain of ApoE. Note that in both panels B and C, the RB domains are colored brown while the side chain moieties of R61, E109, and C112/C112SNO are colored blue, red, and green, respectively. Insets show close-ups of intramolecular interactions of C112/C112SNO with R61 and E109. (D) Schematic showing the S-nitrosylation of C112 within the RB domain of ApoE. Note that the resulting C112SNO S-nitrosothiol derivative may undergo resonance arrangement to form a zwitterion with an internal dipole characterized by the separation of a positive charge and a negative charge on sulfur and oxygen atoms, respectively. (E) Schematic showing a plausible hydrogen bonding and/or ion pairing network of the polarized S-nitrosothiol moiety of C112SNO, the guanidino group of R61, and the side chain carboxylate of E109. The double-headed red arrows indicate potential hydrogen bonding and/or ion pairing contacts. (from reference 14)