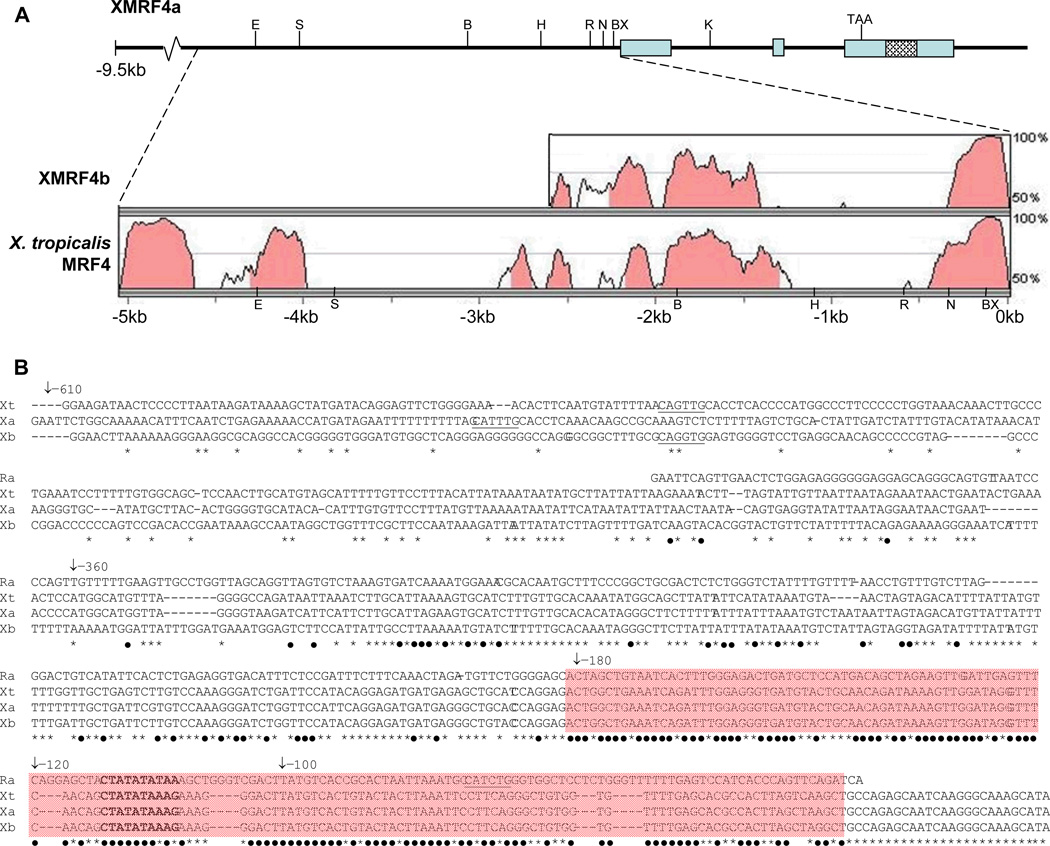
Figure 1. Structure of the XMRF4a genomic clone and comparison to other MRF4 gene sequences.
(A) Upper portion of the figure shows the 5′-flanking region, exons, introns, and 3′-flanking sequence of the 13-kb clone. Restriction enzyme sites mentioned in the text (B, BamH I; BX, BstX I; E, EcoR V; H, Hind III; K, Kpn I; N, Nco I; R, EcoR I; S, Sac I) and the stop codon are indicated. Cross-hatching identifies the P2X7 gene homology region (see text). Lower portion shows results of VISTA alignments of 5066 bp of XMRF4a 5′-flanking sequence versus XMRF4b and X. tropicalis MRF4 loci. Shading indicates conservation of 70% or greater. For XMRF4b, only 2325 bp of sequence was available for comparison. (B) Alignment of nucleotide sequences from rat MRF4 (Ra), X. tropicalis MRF4 (Xt), XMRF4a (Xa), and XMRF4b (Xb) promoters. Numbering refers to positions 5′ from the XlMRF4a start codon. −610 indicates the EcoR I site, −360 the Nco I site, −180 the BstX I site, and −120 and −100 indicate the 5′ ends of PCR-generated deletions. The MEF2 site is in bold type and E boxes (potential MRF binding sites) are underlined. Black dots beneath nucleotides indicate conservation in all four species, while asterisks indicate conservation only among the three Xenopus genes. Shading covers the 150-bp highly conserved region described in the text.