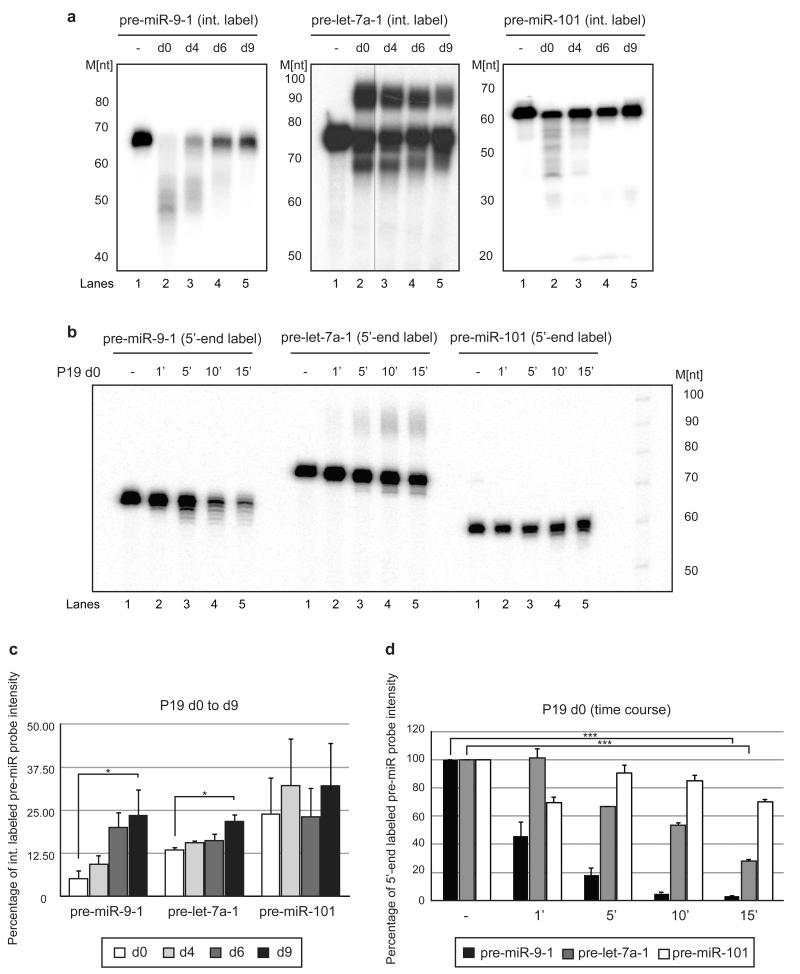
Figure 7. Pre-miR-9 is destabilized in P19 cell extracts from the early stages of neuronal differentiation.
(a) In vitro processing assays were performed with internally radiolabeled pre-miR transcripts (50 × 103 c.p.m. (counts per minute), approximately 20 pmol) in the presence of d0 (Lanes 2), d4 (Lanes 3), d6 (Lanes 4) or d9 (Lanes 4) P19 cell extracts. (-) represents an untreated control. Reactions were supplemented with 0.25 mM UTP. The products were analyzed on an 8% denaturing polyacrylamide gel. [M] - RNA size marker. (b) In vitro processing assays were performed with 5′-end labeled pre-miR transcripts (50 × 103 c.p.m. (counts per minute), approximately 20 pmol). Pre-microRNAs were incubated in d0 P19 cell extracts for 1 (1′), 5 (5′), 10 (10′) or 15 min (15′). The control (-) was incubated without extract for 15′. [M] - RNA size marker. (c) The percentage of pre-microRNA substrate intensity remaining after corresponding in vitro processing reactions was plotted relative to the control reactions set to 100. Mean values and standard deviations (SD) of three independent experiments are shown. Statistical significance was calculated using a t-test (*) −P ≤ 0.05, (***) −P ≤ 0.001.