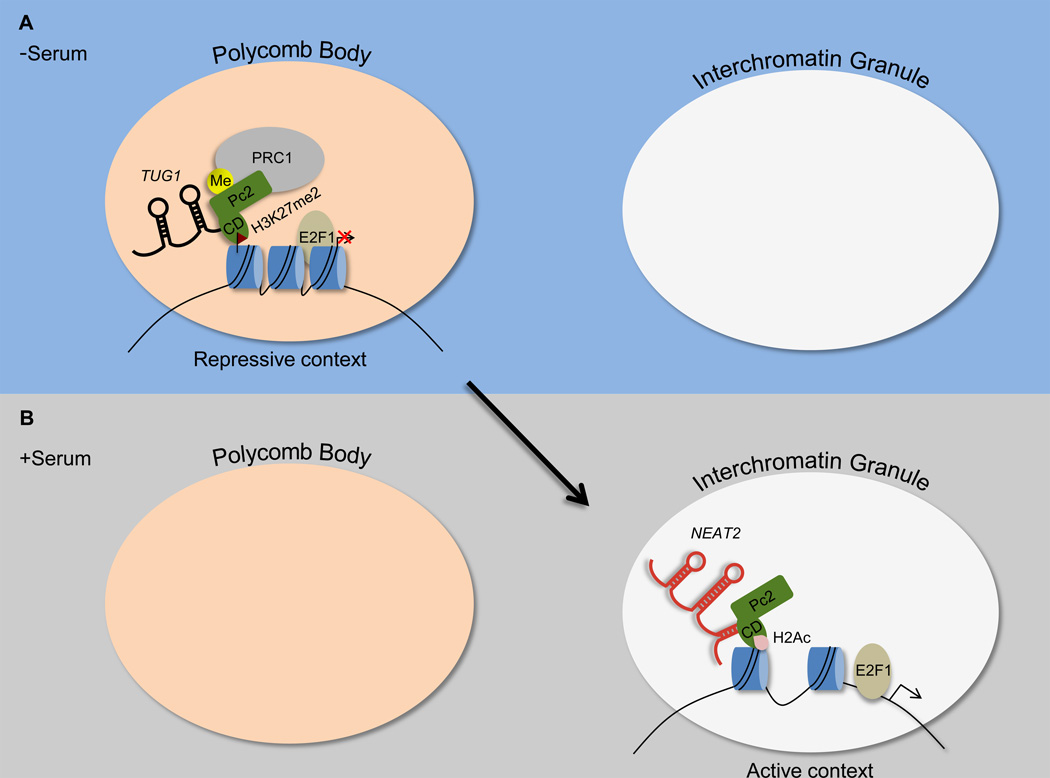
Figure 1. E2F1-responsive genes localize to a Polycomb body or interchromatin granule structure in the absence or presence of serum, respectively, dependent on the methylation status of Pc2.
(A) Under serum starvation conditions, Pc2 is methylated and interacts with TUG1 through its chromodomain (CD), which results in a higher affinity toward binding of H3K27me2. As a result, E2F1-responsive genes localize to a Polycomb body, which provides an environment to reinforce transcriptional repression.
(B) After serum stimulation, Pc2 is demethylated and interacts with NEAT2, which promotes association of the Pc2 CD with acetylated histone H2. As a result, E2F1-responsive genes localize to an interchromatin granule, which harbors a transcriptionally permissive environment.