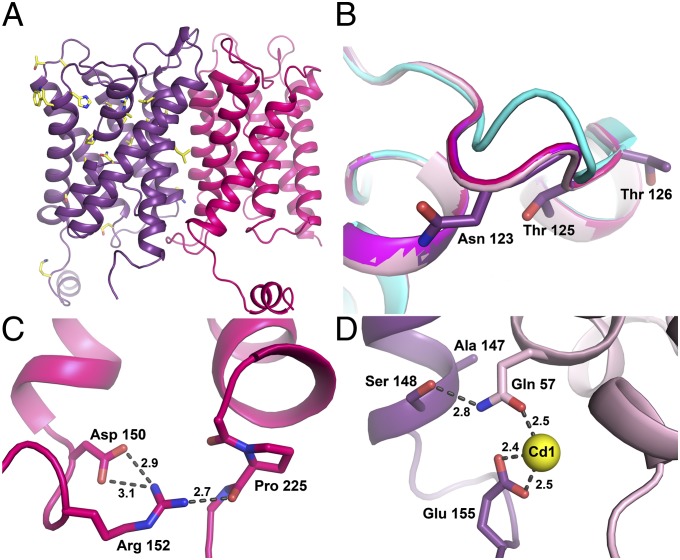
Fig. 4.
NDI-causing mutations in AQP2. (A) Overview of known NDI-causing mutations (yellow) in the AQP2 structure showing how these mostly affect the pore-forming regions. (B) Structural overlay of loop C from the four AQP2s showing the N-glycosylation site at Asn123. A comparison with other mammalian AQPs, here represented by AQP5 (cyan), shows that structure of AQP2 is markedly different at this site. Asn123 as well as two nearby NDI mutation sites, Thr125 and Thr126, are shown in stick representation. (C) Structure around the NDI mutation site at Asp150, showing its connections to Pro225 at the proximal end of the C-terminal tail via Arg152. Hydrogen bonds are indicated by dotted lines. Distances are shown in Å. (D) Close-up of the Cd1 site showing how Ser148 hydrogen bonds to the Cd2+ ligand Gln57. Mutation of both these residues cause ER retention of AQP2. Bonds are indicated by dotted lines with distances shown in Å.