Fig. 1.
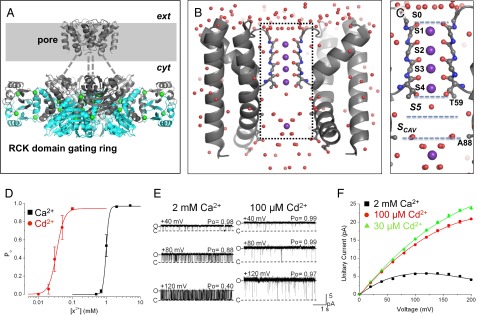
Structure and activation properties of MthK. (A) Presumed biological structure of MthK shown as a Cα-trace [Protein Data Bank (PDB) ID code 3RBZ]. The channel consists of a transmembrane pore domain tethered to a ring of RCK domains, which mediate channel activation by cytoplasmic Ca2+ (green spheres). The gray-shaded region represents the presumed plasma membrane; dashed lines represent the linker region between the pore and RCK gating ring that is unresolved in the crystal structure. (B) High-resolution structure of the MthK pore domain, with the selectivity filter shown in ball and stick representation (PDB ID code 3LDC). Subunits in the front and back have been removed for clear visualization of the conduction pathway (inside dashed rectangle), with K+ ions shown as purple spheres and ordered water molecules shown as red spheres. (C) –Magnified view of the MthK conduction pathway (boxed region in B) with potential ion binding sites (S0–Scav) indicated. (D) Po vs. [Ca2+] (black symbols) and [Cd2+] (red symbols) from currents recorded at −100 mV. MthK activation requires ∼20-fold lower [Cd2+] compared with [Ca2+]. Curves represent fits with a Hill equation with the following parameters: EC50 = 1.0 mM and nH = 9.5 for Ca2+; EC50 = 49 μM and nH = 8.4 for Cd2+. (E) Representative single channel currents from reconstituted MthK at depolarized voltages with 200 mM KCl at both sides of the membrane and Ca2+ or Cd2+ at the cytoplasmic side of the channel as indicated. Cd2+ can fully activate MthK at concentrations that produce much less fast blockade than Ca2+. O and C indicate open and closed current levels, respectively. (F) Unitary current vs. voltage for MthK channels activated with 30 and 100 μM Cd2+ (green and red, respectively) and 2 mM Ca2+ (black). Smooth curves are drawn for display only; 100 μM Cd2+ results in nominal levels of fast blockade, yielding large outward current.
