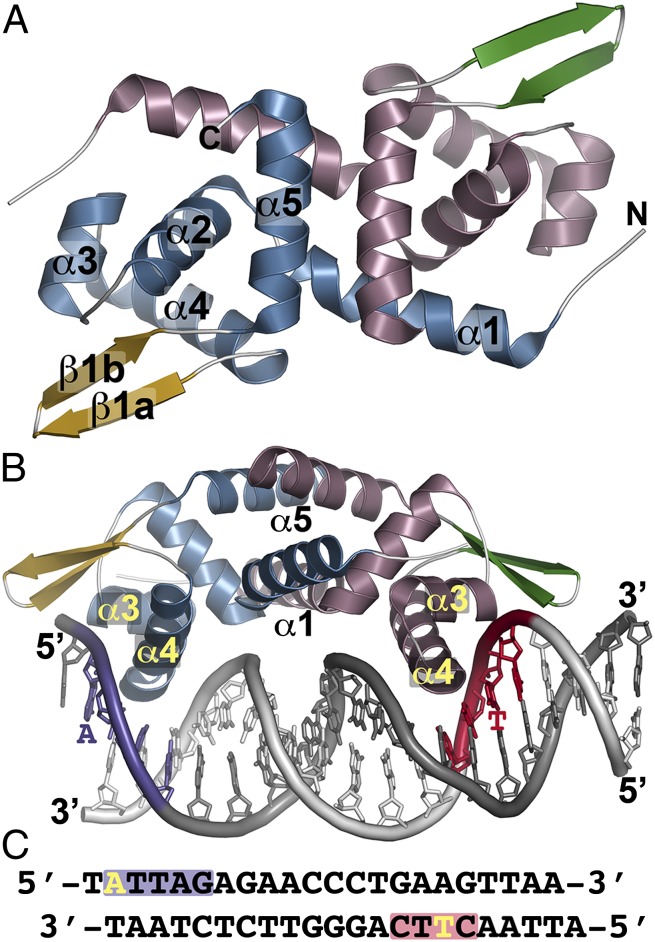
Fig. 1.
Overall structure of NolR. (A) The structure of unliganded NolR is shown as a ribbon diagram. Secondary structure features are labeled in monomer A and are differentially colored in each monomer as follows: blue α-helices and gold β-strands in monomer A and rose α-helices and green β-strands in monomer b. This is a “top” view of the dimeric structure. (B) Structure of NolR in complex with the 22-bp oligo AT duplex. Secondary structure features are colored as in A. The view is rotated ∼90° relative to A to present a “side” view of interaction with DNA. α-Helices forming the dimer interface (α1 and α5) and the helix-turn-helix motif (α3–α4) are labeled. Consensus-motif regions of oligo AT that contact NolR are colored purple and red with key nucleotides indicated. (C) Sequence of oligo AT. The purple and red boxes correspond to the regions of the consensus motif highlighted in B. The yellow A and T indicate nucleotides that are variable in the target DNA sequences of NolR.