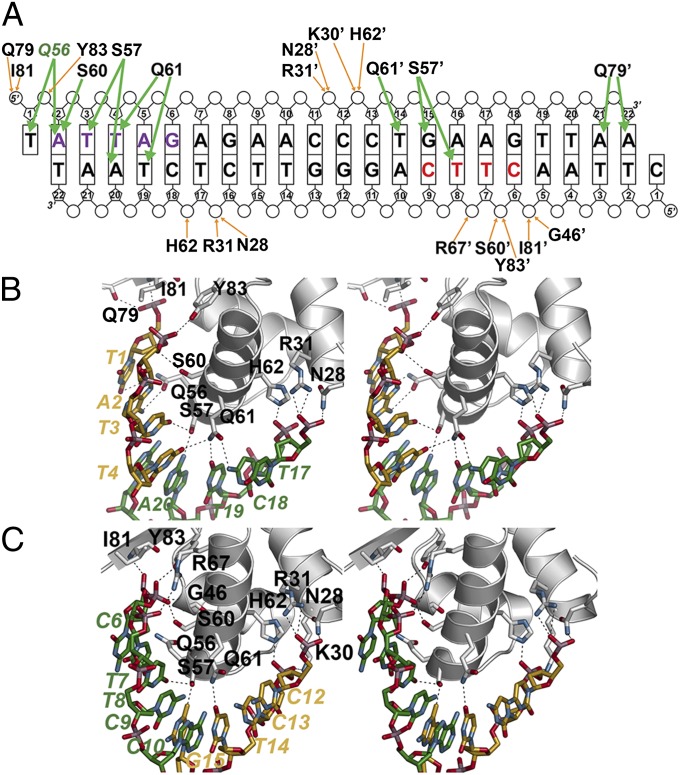
Fig. 2.
NolR and asymmetric operator binding. (A) Schematic showing NolR-oligo AT DNA contacts. The bases are labeled and shown as rectangles, with phosphate and ribose groups drawn as circles and pentagons, respectively. Residues from chain B of the homodimer are noted with an apostrophe after the amino acid number. Orange arrows indicate backbone contacts, and green arrows show base-specific interactions. The two halves of the NolR consensus target sequence that interact with NolR are highlighted with purple and red color, as in Fig. 1 B and C. Gln56 is colored green to emphasize its role in consensus-motif recognition. (B) Stereoview of protein–DNA interactions in the first half-site. Protein side-chains are from chain A. Nucleotides from 5′ and 3′ strands are colored gold and green, respectively, and are labeled. (C) Stereoview of protein–DNA interactions in the second half-site. Protein side-chains are from chain B. Nucleotides from 5′ and 3′ strands are colored gold and green, respectively, and are labeled.