Fig. 3.
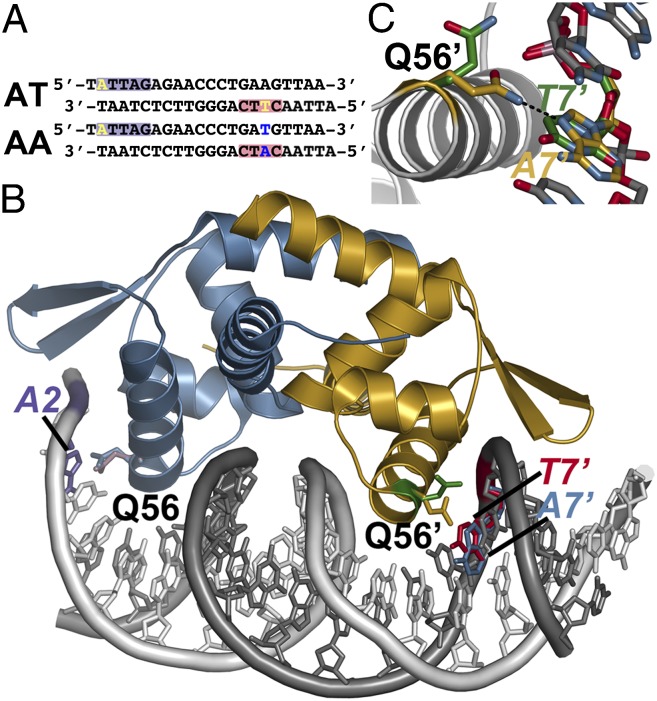
Conformational switching of Gln56 for recognition of variable DNA target sites by NolR. (A) Sequences of oligos AT and AA. The purple and red boxes correspond to the regions of the consensus motif highlighted in Fig. 1 B and C. The variable position bases are highlighted yellow in oligo AT. Changes at these positions in oligo AA are highlighted in blue. (B) Structure of NolR in complex with the 22-bp oligo AA duplex. Chains A and B of NolR are colored blue and gold, respectively. The 5′ and 3′ strands of oligo AA are colored white and gray, respectively. The orientation of the Gln56 side-chain in each monomer of NolR complexed with either oligo AT (purple and green sticks) or oligo AA (blue and gold sticks) is shown. The positions of A2 (purple) from oligo AA, T7' (red) from oligo AT, and A7' (blue) from oligo AA are shown. (C) Close-up view of Gln56 movement in the second consensus half-site of NolR. The position of Gln56 of chain B and the variable base is shown. The structures observed with NolR complexed with either oligo AT (green) and oligo AA (gold) are shown.