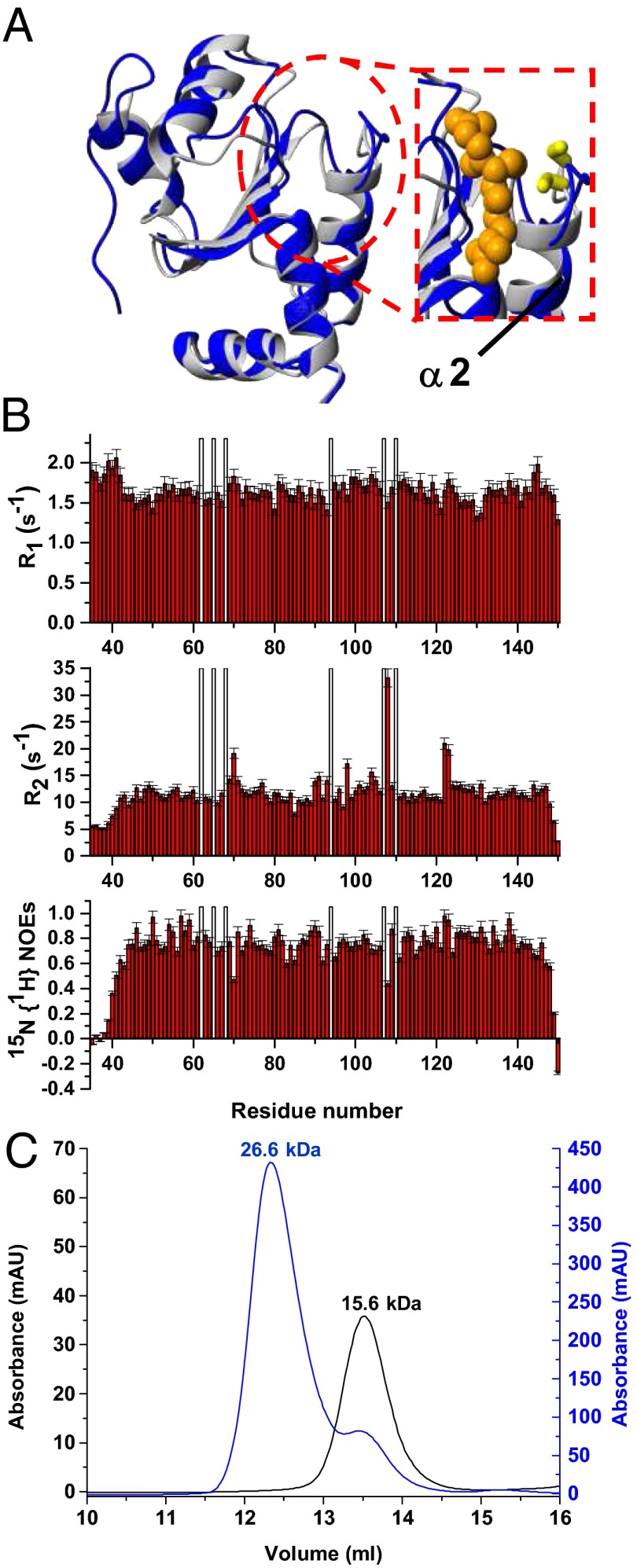
Fig. 4.
Solution structure and dynamics of apo hGRX5 and effects on the aggregation state of hGRX5 induced by [2Fe-2S] cluster binding. (A) Overlay of the solution NMR structure of apo hGRX5 (blue) with a subunit of the tetrameric crystal structure of holo hGRX5 (gray). A zoom of the GSH binding site is shown on the right, where the side chain of the iron binding cysteine and the atoms of glutathione molecule (obtained from the crystal structure) are shown as yellow sticks and orange spheres, respectively. (B) 15N relaxation parameters R1, R2, 15N{1H} NOE versus residue number of apo hGRX5 obtained at 600 MHz and 298 K. The protein was in 50 mM phosphate buffer (pH 7.0) containing 5 mM DTT, 5 mM GSH, and 10% (vol/vol) D2O. White bars indicate Pro residues and the unassigned backbone NH of Gly-68. (C) [2Fe-2S] cluster binding results in a change of the quaternary structure of hGRX5. Analytical gel filtration of apo hGRX5 (black, left y axis) and of holo hGRX5 (blue, right y axis) in degassed 50 mM potassium phosphate buffer (pH 7.0) containing 5 mM DTT and 5 mM GSH. All prepared holo hGRX5 samples contained a residual amount of apo form (ranging from 10 to 30%) that was not possible to remove by gel filtration column in the last purification step (see SI Text for details). Apparent molecular weights are reported on the top of the peaks according to the markers run under the same conditions. The molecular weight of apo hGRX5, as obtained by MALDI-MS and electrospray ionization-MS data, is 13,158 Da, in agreement with the theoretical value (13,157.9 Da) for a monomeric protein state.