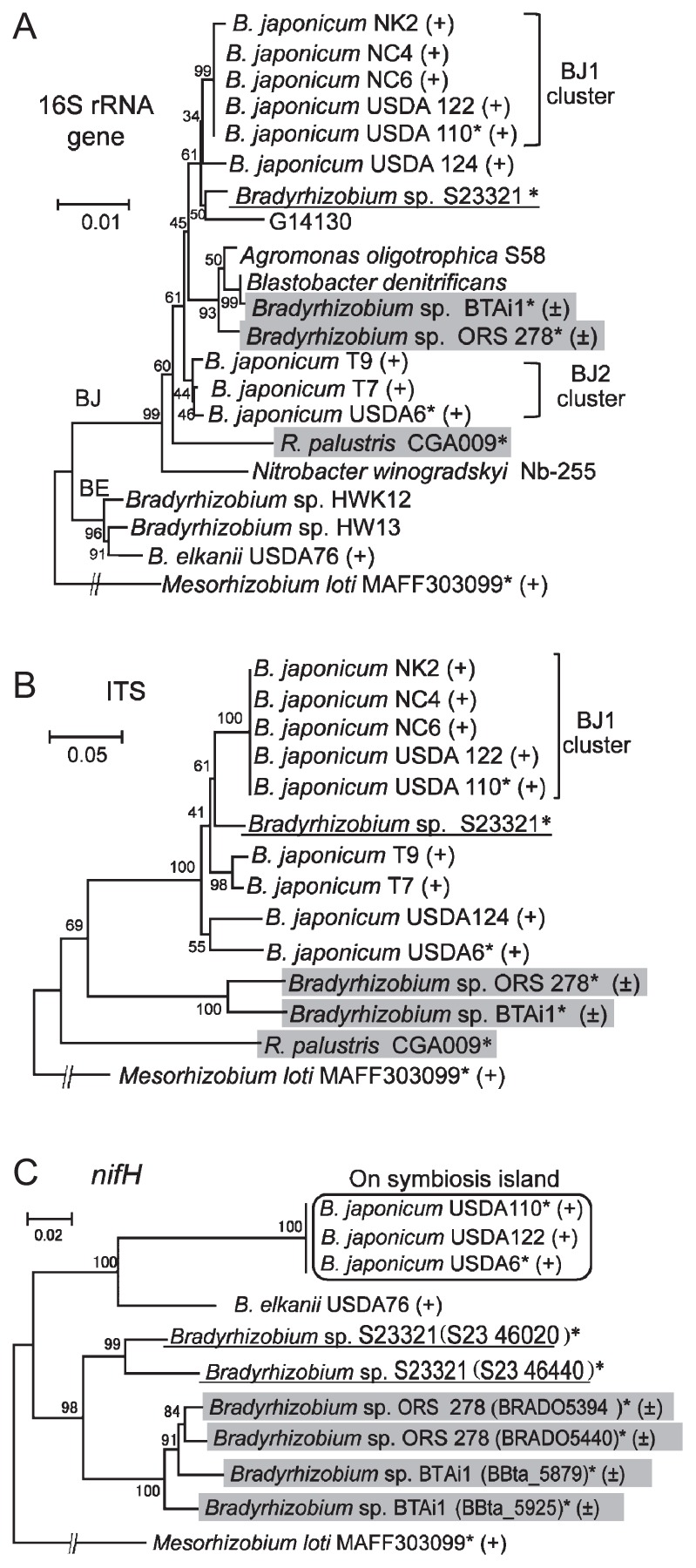
Fig. 1.
Phylogenetic relationships of Bradyrhizobium sp. S23321 and other members of the Bradyrhizobiaceae (Table S1) based on 16S rRNA gene sequences (A), internal transcribed spacer (ITS) sequences (B), and nifH gene sequences (C). For all trees, Mesorhizobium loti MAFF303099 was used as an outgroup. Numbers at the nodes are the percentage of 1,000 bootstrap replications supporting that partition. Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates are collapsed. BJ and BE are major clusters including B. japonicum and B. elkanii, respectively. The BJ1 and BJ2 clusters of B. japonicum were defined based on phylogenetic trees of 16S rRNA genes and ITS sequences as described previously (26). Strains capable of Nod factor-dependent and -independent nodulation (15) are marked with (+) and (±), respectively. Photosynthetic strains are shaded in gray. Asterisks show the strains of bradyrhizobia for which the complete genome sequence is available. S23321 is underlined. In the phylogenic trees based on 16S rRNA gene sequences (A) and ITS sequences (B), S23321 was clustered with B. japonicum. In the nifH tree (C), S23321 was closer to Bradyrhizobium sp. BTAi1 and ORS278 than to the nif genes on the symbiosis islands of B. japonicum (rectangle in panel C).