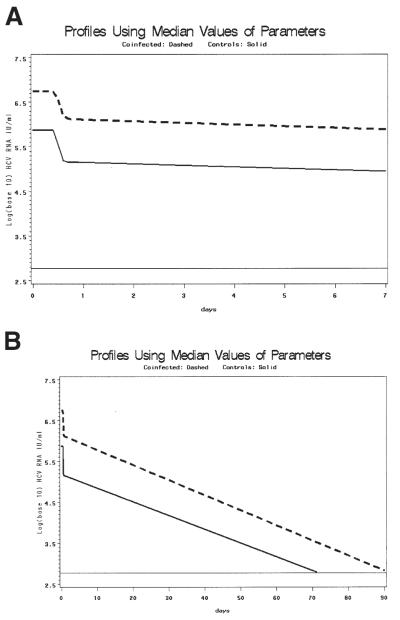
Figure 5.
(A, B) Graphs using median kinetic parameters from genotype 1 patients to show delayed clearance in co-infected patients: all parameters (∈, c, δ, V0, t0) were calculated from the median pooled values of the study population for whom the model converged and who had viral clearance. The estimated V0 was 6.86 log10 IU/mL for co-infected and 5.88 log10 IU/mL for monoinfected patients. T0 was 11.39 vs. 11.26 hours for co-infected and monoinfected patients, respectively; c was 33.20 and 22.67/day, respectively; δ was .11 and .10/day, respectively; and ∈ was .76 and .81, respectively. These parameters were used to calculate λ1 and λ2 and the resulting pooled model fits. Solid line represents co-infected patients, dashed line represents monoinfected patients. The lower limit of assay detection is represented by the thin solid line at log10(600) IU/mL. (A) Early viral clearance (through 7 days). (B) Extension of model estimation through clearance (90 days). There is an approximately 20-day differential for predicted time to clearance between co-infected and monoinfected patients.