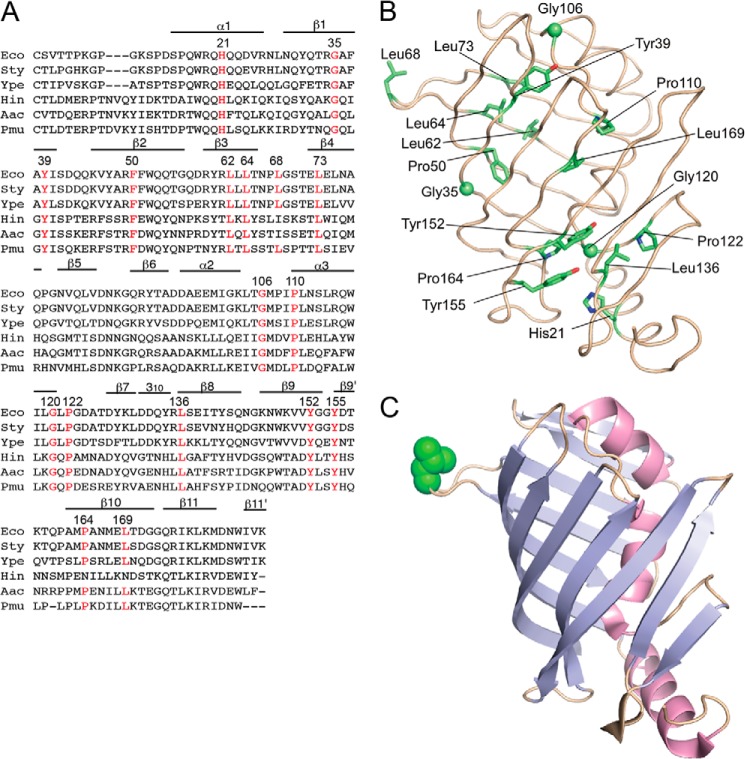
FIGURE 1.
Sequence alignment of LolB homologues and positions of mutated residues. A, the amino acid sequences of LolB and its homologues were aligned using ClustalW (26). Residues forming α and 310 helices and β-strands are indicated. Conserved residues that were subjected to random mutagenesis are indicated in red with their position numbers. Eco, E. coli; Sty, Salmonella enterica serovar typhimurium; Ype, Yersinia pestis; Hin, Haemophilus influenzae; Aac, Actinobacillus actinomycetemcomitans; Pmu, Pasteurella multocida. B, the positions of mutated residues are shown in the structure of mLolB. Side chains of the residues are shown as sticks, whereas Cα atoms of glycine are shown as spheres. C, mLolB is represented as a ribbon model in which strands and helices are colored cyan and red, respectively. The side chain of Leu-68 in the hydrophobic loop is shown as a CPK model in green.