Background: Two Fanconi anemia patient missense mutations are localized in FANCJ helicase motif Ia.
Results: Mutant R251C impairs the DNA binding ability of FANCJ; Q255H uncouples DNA translocation from helicase activity.
Conclusion: Helicase motif Ia plays critical roles in FANCJ enzymatic activity and DNA repair.
Significance: Helicase motif Ia is involved not only in nucleic acid binding but also ATP binding and coupling ATP hydrolysis to unwinding.
Keywords: DNA Helicase, DNA Repair, Enzymes, Mutant, Pathogenesis, FANCJ, Fanconi Anemia, Helicase, Motif Ia
Abstract
Helicases are molecular motors that couple the energy of ATP hydrolysis to the unwinding and remodeling of structured DNA or RNA, which is coordinated by conserved helicase motifs. FANCJ is a DNA helicase that is genetically linked to Fanconi anemia, breast cancer, and ovarian cancer. Here, we characterized two Fanconi anemia patient mutations, R251C and Q255H, that are localized in helicase motif Ia. Our genetic complementation analysis revealed that both the R251C and Q255H alleles failed to rescue cisplatin sensitivity of a FANCJ null cell line as detected by cell survival or γ-H2AX foci formation. Furthermore, our biochemical assays demonstrated that both purified recombinant proteins abolished DNA helicase activity and failed to disrupt the DNA-protein complex. Intriguingly, R251C impaired DNA binding ability to single-strand DNA and double-strand DNA, whereas Q255H retained higher binding activity to these DNA substrates compared with wild-type FANCJ protein. Consequently, R251C abolished its DNA-dependent ATP hydrolysis activity, whereas Q255H retained normal ATPase activity. Physically, R251C had reduced ATP binding ability, whereas Q255H had normal ATP binding ability and could translocate on single-strand DNA. Although both proteins were recruited to damage sites in our laser-activated confocal assays, they lost their DNA repair function, which explains why they exerted a domain negative effect when expressed in a wild-type background. Taken together, our work not only reveals the structural function of helicase motif Ia but also provides the molecular pathology of FANCJ in related diseases.
Introduction
DNA helicases are involved in all aspects of DNA metabolism, including DNA replication, DNA repair, recombination, transcription, and telomere maintenance (1–3). All helicases are endowed with nucleic acid-dependent ATPase activity, but only some have ATP-dependent DNA unwinding activity (4). Although they differ in the amino acid sequence in their extreme N- and C-terminal parts, they all contain a helicase core domain, a region of 200–700 amino acids composed of 7 conserved motifs (I, Ia, II, III, IV, V, and VI) (5). Generally, motifs I, II, and VI are essential for ATP binding and hydrolysis, motifs IV and V are primarily responsible for DNA binding, and motif III is important for coupling ATP hydrolysis to remodeling events such as unwinding (6, 7).
The function of helicase motif Ia has been less well studied, although it is generally accepted that motif Ia functions in substrate DNA or RNA binding. The structures of Rep with single-strand DNA (ssDNA,2 dT15) (8), PcrA with partially double-strand DNA (dsDNA, 10 bp of dsDNA with a 7-nucleotide ssDNA tail) (9), hepatitis C virus (HCV) NS3 helicase with oligo (dU8) (10) and its modeled structure with RNA substrates (11), and Hel308 with a partial dsDNA (15 bp of dsDNA with a 10-base single-stranded 3′ tail) (12) have demonstrated that residues from helicase motif Ia contact nucleic acids. The three crystal structures of archaeal XPD have recently been solved (13–15), all showing that the amino acids of motifs Ia, III, and V contribute to the DNA binding domain. Mutagenesis analysis of motif Ia in the herpes simplex virus type 1 origin-binding protein UL9 revealed that motif Ia is involved in ssDNA binding and helicase activities (16).
Fanconi anemia group J protein (FANCJ), also known as BRIP1, is a superfamily 2 DNA helicase. The identification of FANCJ mutations in early onset breast cancer patients (17, 18), Fanconi anemia (FA) group J patients (19–21), and ovarian cancer patients (22) implicates FANCJ as a tumor suppressor caretaker that functions in DNA interstrand cross-link repair and double-strand break repair (21). FANCJ encodes a 1249-amino acid protein and contains seven conserved helicase motifs (23) and a Q motif that regulates protein dimerization (24). FANCJ has been demonstrated biochemically to be a bona fide DNA helicase that catalytically unwinds duplex DNA (25, 26) and G-quadruplex structures (27, 28) in an ATP hydrolysis-dependent manner. Helicase activity of FANCJ is required for timely progression through S phase (29). FANCJ physically and functionally interacts with human replication protein A (30), mismatch repair complex MutLα (31), Bloom's syndrome helicase (32), and MRE11 nuclease (33). FANCJ binds and acts with TopBP1 in early DNA replication checkpoint control (34), which suggests that FANCJ has additional roles in the response to replication stress (35) and operates in a parallel pathway to the classic FA pathway (36). Several genotyping studies have identified the association between FANCJ mutations and FA clinical abnormalities (19, 20), breast cancer risk (18, 37–42), and ovarian cancer risk (22). Two FANCJ missense mutations, arginine to cysteine at residue 251 (R251C) and glutamine to histidine at 255 (Q255H), have been reported in FA patients (Fanconi Anemia Mutation Database) (19). Interestingly, both disease-causing mutations are localized in helicase motif Ia with a distance of three amino acids between. However, the molecular mechanism by which these two mutations lead to FA remains unknown.
In this study we characterized the biochemical and cellular effects of the FANCJ-R251C and Q255H mutants. Both proteins abolished DNA helicase activity and failed to disrupt the DNA-protein complex. Interestingly, R251C impaired DNA binding ability, whereas Q255H retained higher DNA binding activity to single-strand DNA and double-strand DNA compared with wild-type FANCJ. Consequently, R251C abolished its ATP hydrolysis activity; Q255H contained normal DNA-dependent ATPase activity. However, both the R251C and Q255H alleles failed to rescue cisplatin sensitivity of a FANCJ null cell line as detected by cell survival or γ-H2AX foci formation. Furthermore, both mutant alleles exerted a domain negative effect when expressed in a wild-type background. These results suggest that FANCJ helicase activity, but not ATPase or translocase activity, is required for FANCJ function in DNA repair.
EXPERIMENTAL PROCEDURES
Plasmid DNA Construction
The FANCJ-R251C and FANCJ-Q255H mutations were generated by the Stratagene QuikChange site-directed mutagenesis kit according to the manufacturer's instructions using respective primers (supplemental Table S1) in the vectors of pVL1392 (25) and pEGFP-C2 (43) that contain the wild-type FANCJ gene. All desired mutations and constructs were confirmed by direct DNA sequencing using the purified plasmids.
Recombinant Proteins
Baculovirus encoding FANCJ-wild type (WT), -R251C, or -Q255H with a C-terminal FLAG tag was used to infect High Five insect cells, and the recombinant FANCJ proteins were purified with modifications to a protocol previously described (28, 43). Briefly, cell pellets were resuspended in buffer A (10 mm Tris·HCl (pH 7.5), 130 mm NaCl, 1% Triton X-100, 10 mm NaF, 10 mm NaPi, 10 mm NaPPi). Cells were lysed in the presence of protease inhibitors (Roche Applied Science) for 45 min at 4 °C with mild agitation and centrifuged at 21,000 × g for 10 min at 4 °C. The supernatant was incubated with FLAG antibody resin (Sigma) for 2 h at 4 °C. The resin was washed twice with buffer B (50 mm Tris·HCl (pH 7.4), 500 mm NaCl, and 0.5% Nonidet P-40) followed by buffer C (50 mm Tris·HCl (pH 7.4), 150 mm NaCl, 0.5% Nonidet P-40). FANCJ was eluted with 4 μg/ml 3× FLAG peptide (Sigma) in buffer D (25 mm Tris·HCl (pH 7.4), 100 mm NaCl, 10% glycerol, 0.1% Tween 20, 5 mm Tris (2-carboxyethyl) phosphine hydrochloride; Sigma) for 1 h. FLAG-tagged FANCJ proteins were dialyzed for 2 h against buffer D using a dialysis tube with a 50-kDa molecular mass cut-off (Tube-O-DIALYZER™), and aliquots were frozen in liquid nitrogen and stored at −80 °C. Protein concentration was determined by the Bradford assay using BSA as a standard.
DNA Substrates
Polyacrylamide gel electrophoresis purified oligonucleotides used for the preparation of DNA substrates were purchased from IDT (Integrated DNA Technologies) and are listed in supplemental Table S1. The forked duplex DNA substrate was prepared from the DC26 and TSTEM25 oligonucleotides, and the G4 DNA substrate was prepared from oligonucleotide TP as previously described (28).
Helicase Assays
Helicase assay reaction mixtures (20 μl) contained 40 mm Tris-HCl (pH 7.4), 25 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 2% glycerol, 100 ng/μl bovine serum albumin, 2 mm ATP, 0.5 nm concentrations of the specified tetraplex or duplex DNA substrate, and the indicated concentrations of FANCJ. Helicase reactions were initiated by the addition of FANCJ and incubated at 30 °C for 15 min unless otherwise indicated. Helicase reactions were terminated by addition of Stop buffer containing EDTA. Reaction products were resolved on non-denaturing 8% or 12% (19:1 acrylamide:bisacrylamide) polyacrylamide gels for the G4 and forked duplex substrates, respectively. The resolved radiolabeled species were visualized with Typhoon 9400 phosphorimaging and analyzed with ImageQuant 5.2 software (GE Healthcare).
Streptavidin Displacement Assays
Streptavidin displacement reaction mixtures (20 μl) contained 40 mm Tris-HCl (pH 7.6), 25 mm KCl, 2 mm MgCl2, 2% glycerol, 100 ng/μl bovine serum albumin, 2 mm ATP, 0.5 nm concentrations of streptavidin-bound biotinylated oligonucleotide (X12-1-52-BIOT13, supplemental Table S1), and the indicated concentrations of FANCJ helicase. For streptavidin displacement reactions, 0.5 nm of biotinylated oligonucleotide was preincubated with 100 nm streptavidin (Sigma) for 10 min at 37 °C. Streptavidin bound to the DNA substrate was detected by a slower migrating gel-shifted species on nondenaturing 12% polyacrylamide gel. Reactions were initiated by adding FANCJ before the addition of biotin (1 μm) and incubated at 30 °C for 15 min followed by a quench with the addition of 10 μl of stop buffer (50 mm EDTA, 40% glycerol, 0.9% SDS, 0.05% bromphenol blue, and 0.05% xylene cyanol). Products were resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) polyacrylamide gels. The resolved radiolabeled species were visualized and analyzed as described above.
Electrophoretic Mobility Shift Assay
Protein/DNA binding mixtures (20 μl) contained the indicated concentrations of FANCJ and 0.5 nm concentrations of the specified 32P-end-labeled DNA substrate in the same reaction buffer as used for helicase assays (see above) containing 2 mm ATPγS or no nucleotide. The binding mixtures were incubated at room temperature for 30 min after the addition of FANCJ. After incubation, 3 μl of loading dye (74% glycerol, 0.01% xylene cyanol, 0.01% bromphenol blue) were added to each mixture, and samples were loaded onto native 5% (19:1 acrylamide:bisacrylamide) polyacrylamide gels and electrophoresed at 200 V for 2 h at 4 °C using 1× Tris borate-EDTA as the running buffer. The resolved radiolabeled species were visualized and analyzed as described above.
ATP Hydrolysis Assays
ATP hydrolysis was measured using [γ-32P] ATP (PerkinElmer Life Sciences) and analyzed by thin-layer chromatography (TLC) on polyethyleneimine-cellulose plates (Mallinckrodt Baker). The standard reaction mixture (20 μl of total volume) contained 40 mm Tris-HCl (pH 7.4), 25 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 2% glycerol, 100 ng/μl bovine serum albumin, 250 μm [γ-32P] ATP, and 60 nm FANCJ protein and was incubated at 30 °C. The product was spotted onto a PEI-cellulose TLC plate and resolved by using 0.5 m LiCl and 1 m formic acid as the carrier solvent. The TLC plate was exposed to a phosphorimaging screen for 1 h, and the radiolabeled species were visualized and analyzed as described above.
ATP Binding Assays
ATP binding assays (30 μl) were performed in the same reaction buffer as for helicase or ATPase assays as described above with 5 μCi [α-32P]ATP (3000 Ci/mmol, PerkinElmer Life Sciences). Assays were initiated by adding FANCJ protein to a final concentration of 230 nm followed by incubating at 4 °C for 30 min. Reactions were then applied to Bio-Spin P30 Tris chromatography columns (Bio-Rad) that had been pre-equilibrated in a reaction buffer. One-drop (∼45 μl) fractions were collected as flow-through under gravity from a column that was eluted with Tris-EDTA. The specific radio activity of each fraction was determined by a liquid scintillation counter (MicroBeta TriLux, PerkinElmer Life Sciences). The first peak (3–4 drops) was considered to be protein-bound ATP, and the second peak was considered to be unbound ATP.
Fluorescence Quench Translocation Assays
High pressure liquid chromatography-purified, fluorescein-labeled oligonucleotide was purchased from Loftstrand Labs (Gaithersburg, MD) with the fluorophore attached at the 3′ end (T50F, supplemental Table S1). Translocation assays were performed in 96-well F-bottom plates (Greiner Bio-One) using 1 nm fluorescein-labeled probe and 19.2 nm FANCJ protein in 50-μl reaction mixtures using the ATPase reaction buffer described above with 2 mm ATP. Translocase assays were initiated with the addition of FANCJ protein and incubated at room temperature (22 °C) with the specified reading point. Fluorescence readings were taken on a FLUOstar plate reader (BMG LabTech) with excitation and emission filters at 485 and 520 nm, respectively. Background fluorescence (<10%) in reactions at time 0 was subtracted from the fluorescence signals detected in time course reactions.
Size Exclusion Chromatography
Size exclusion chromatography analysis of FANCJ proteins was performed as previously described (24). Briefly, purified recombinant FANCJ protein (0.2 mg/ml, 0.5 ml) was applied to a 24-ml Superdex-200 size exclusion column (GE Healthcare) using an AKTA FPLC (GE Healthcare) that was equilibrated and eluted with 25 mm Tris·HCl (pH 7.5), 10% glycerol, 0.15 m NaCl, 1 mm EDTA, and 0.5 mm DTT. The column was run at a rate of 0.1 ml/min, and 0.5-ml fractions were collected. Proteins were detected using a UV detector. Protein concentration was determined by the Bradford method using BSA as a standard in a 96-well plate. Enzymatic activity of the FANCJ protein was examined with the standard protocol as described above. The column was calibrated using standard molecular mass markers containing blue dextran (2000 kDa), thyroglobulin (670 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and aprotinin (6.5 kDa) (Sigma).
Cell Lines
U2OS and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine at 37 °C in 5% CO2. For transient transfection, Lipofectamine2000 (Invitrogen) was used according to the provider's protocol, and cells were used 48 h after transfection.
Chicken DT40 cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum, 1% (v/v) chicken serum, 2 mm l-glutamine, 50 μm 2-mercaptoethanol, and 1% (v/v) penicillin-streptomycin in a 5% CO2 incubator at 41 °C. Generation of fancj knock-out cell lines has been described elsewhere (43). The fancj cell lines were transfected with plasmids encoding green fluorescence protein (GFP), GFP-FANCJ-WT, GFP-FANCJ-R251C, and GFP-FANCJ-Q255H and selected by G418 resistance using a previously described procedure (43). Colony formation assays were carried out in medium containing 1.4% (w/v) methylcellulose and cisplatin at the dosages previously described (43).
Laser Irradiation and Confocal Microscopy
Immunofluorescence localization of FANCJ proteins at laser-activated psoralen-ICLs (Pso-ICLs) or laser-induced DSBs in vivo was performed as described previously (33) with the following modifications. Briefly, a Nikon Eclipse 2000E spinning disk confocal microscope with five laser imaging modules and a charge-coupled device (CCD) camera (Hamamatsu) was employed. The setup integrated a Stanford Research Systems (SRS) NL100 nitrogen laser with a Micropoint ablation system (Photonics Instruments). Site-specific DNA damage was induced using the SRS NL100 nitrogen laser adjusted to emit at 365 nm. Positions internal to the nuclei of either live untransfected cells or cells transfected with GFP-tagged FANCJ plasmids were targeted using a 60× oil objective lens. Cells were targeted at 5.5% laser intensity to induce DSBs or 1.7% laser intensity to create Pso-ICLs, and images were captured at various time points and analyzed using Volocity, Version 5.0, build 6 (Improvision). The exposure to the laser at the intensities employed in these experiments did not affect cell viability as assayed 24 h after treatment. Experiments were performed using an environmental chamber attached to the microscope to maintain experimental conditions (i.e. 37 °C, 5% CO2, and 80% humidity). At the indicated time intervals, cells from different areas of the dish were targeted with the laser to generate a time course on a single plate. After the final time point, cells were fixed immediately in freshly prepared 4% formaldehyde in PBS for 15 min at room temperature.
Immunofluorescence Studies
Cells were fixed with freshly prepared 4% formaldehyde at room temperature for 15 min. Fixed cells were washed 4 times with PBS and treated with 0.5% Triton X-100 solution (Sigma) at room temperature for 10 min. Cells were washed 4 times with PBS and blocked with 10% goat serum (Sigma) overnight at 4 °C. Indirect immunostaining was performed by first incubating cells with a mouse anti-γ-H2AX monoclonal antibody (1:500, Millipore) overnight at 4 °C. After 4 washes in PBS with 0.1% Tween 20, cells were incubated with Alexa Fluor 488 goat anti-mouse IgG (1:400, Invitrogen) for 1 h at room temperature. Cells were washed 4 times with PBS containing 0.1% Tween 20 and coated with Prolong Gold anti-fade reagent containing DAPI (Invitrogen). Coverslips were placed on chamber slides, and cells were cured at room temperature in the dark for 24 h. Immunofluorescence analyses were performed with a Zeiss LSM 510 META inverted Axiovert 200M laser scan microscope with a Plan-Apochromat 63 × 1.4-numerical-aperature oil immersion differential interference contrast objective lens. Images were captured with a CCD camera and analyzed using LSM Browser software (Zeiss).
Statistical Analyses
Statistical significance was calculated using Student's t test by assuming a two-sample equal variance with a two-tailed distribution.
RESULTS
FANCJ-R251C and FANCJ-Q255H Fail to Complement FANCJ Deficiency
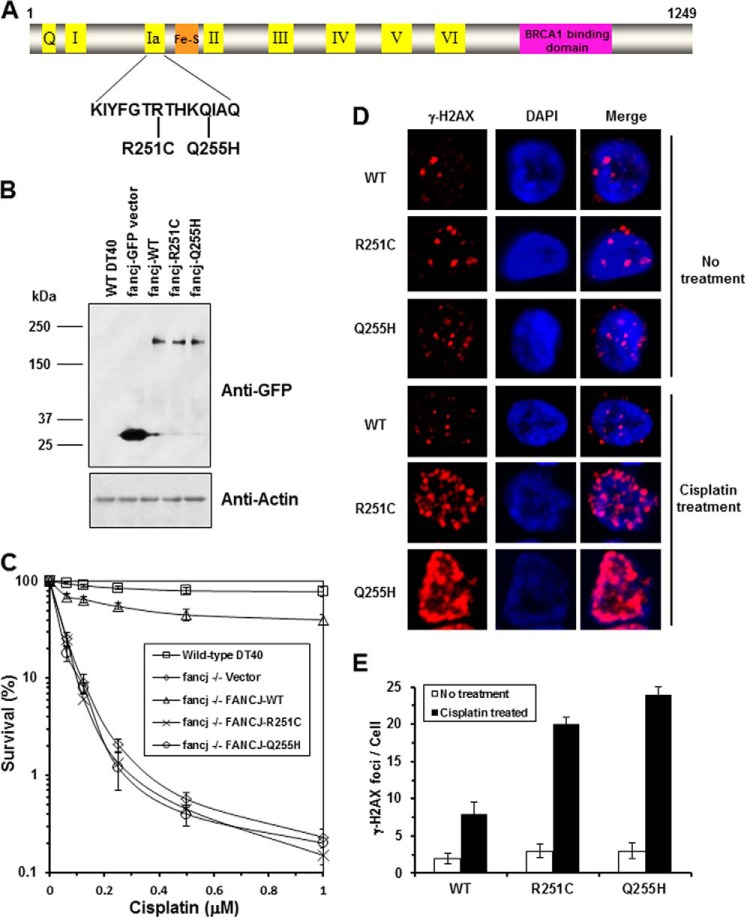
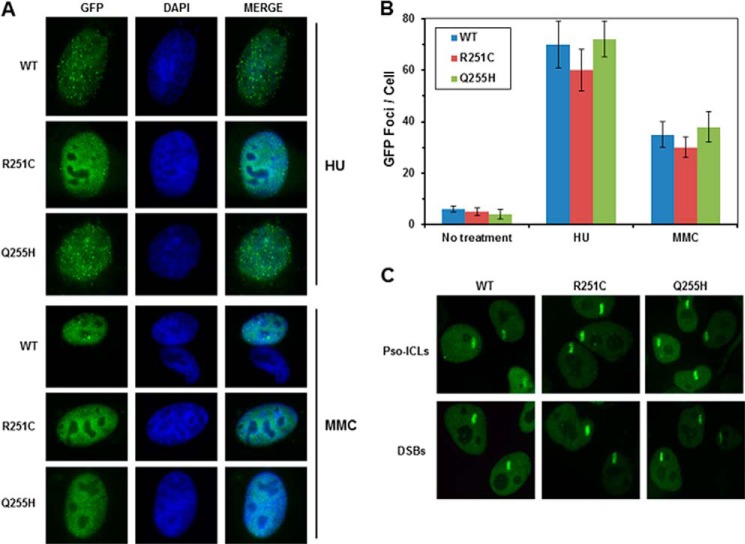
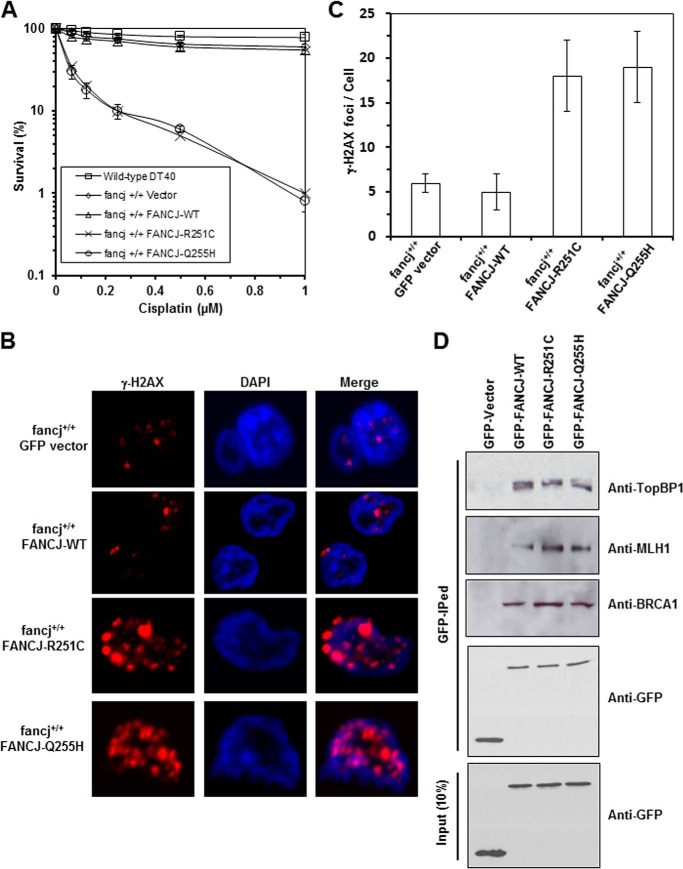
Two FANCJ polymorphisms, R251C and Q255H, have been reported in FA patients. Interestingly, these two mutations are localized in FANCJ helicase motif Ia separated by a distance of three amino acids (Fig. 1A). To determine whether the R251C and Q255H mutants could function in vivo, we performed a genetic complementation experiment using a chicken DT40 cell line with a deletion mutation in the fancj gene. Because the genetic functions of chicken and human FANCJ are conserved (24, 43, 44), we were able to perform genetic assays with the fancj null cell line that had been transfected with plasmids encoding human FANCJ-WT, -R251C, or -Q255H proteins. Western blot analysis demonstrated that R251C and Q255H were expressed at a similar level as the WT (Fig. 1B). The results of cisplatin survival assays, as measured by a colony formation assay, showed that the fancj−/− R251C and fancj−/− Q255H cells were sensitive to cisplatin (Fig. 1C). Compared with the fancj−/− WT cells, fancj null cells and fancj null cells transfected with either empty vector or R251C or Q255H showed very similar reductions in percent survival as a function of cisplatin dose and were ∼250-fold more sensitive to the highest concentration of cisplatin (1 μm). γ-H2AX foci, a marker of double-strand break (DSB), was significantly (3-fold) elevated in cisplatin-treated fancj−/− R251C or fancj−/− Q255H cells compared with the isogenic fancj−/− WT cells (Fig. 1, D and E). These results indicated that FANCJ-R251C and FANCJ-Q255H failed to render the cells resistant to the effects of the cross-linking agent, as measured by cell viability and the accumulation of double strand breaks.
FIGURE 1.
Expression of FANCJ-R251C or FANCJ-Q255H fails to rescue sensitivity of fancj mutant cells to the DNA cross-linker cisplatin. Panel A, schematic depicting FANCJ protein with the conserved helicase core domain and position of the FA causing mutations in motif Ia. The Fe-S domain and BRCA1 binding domain are indicated. Panel B, Western blot analysis of wild-type DT40 cells or fancj null cells transfected with plasmids encoding GFP, GFP-FANCJ-WT, GFP-FANCJ-R251C, or GFP-FANCJ-Q255H. Protein was detected with antibody against GFP (1:1000, Clontech) or actin (as a loading control, 10% loaded). Panel C, cisplatin sensitivity of cells with the indicated genotypes was evaluated by colony formation assay. Panel D, γ-H2AX immunofluorescence staining of fancj null cells transfected with plasmids encoding FANCJ-WT or FANCJ-R251C or FANCJ-Q255H. DT40 cells were treated with or without 1 μm cisplatin for 12 h followed by immunofluorescence detection as described under “Experimental Procedures.” Panel E, quantitative analyses of γ-H2AX foci in the corresponding transfected fancj null cell lines shown in panel D. Data represent the mean of at least 100 cells counted, with the S.D. indicated by error bars. Using a Student's t test for analysis of the γ-H2AX foci data (cisplatin treated), the differences in p values between fancj−/− FANCJ-WT cells and fancj−/− FANCJ-R251C or fancj−/− FANCJ-Q255H were 0.0002 and 0.00005, respectively, indicating a significant difference (p < 0.01) for each.
Both FANCJ-R251C and FANCJ-Q255H Mutants Abolish DNA Helicase Activity
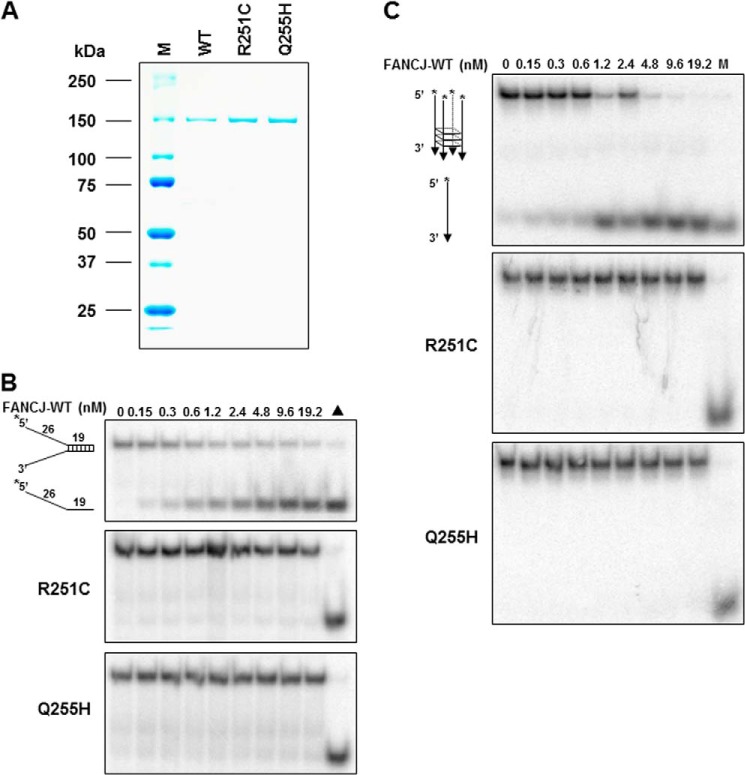
To examine the molecular defects of DNA repair in vivo, we investigated the biochemical character of these two mutants. The recombinant proteins of human FANCJ-WT, -R251C, and -Q255H were expressed using a baculovirus system and purified in an identical manner. Three proteins were purified to near homogeneity as demonstrated by single Coomassie-stained bands on SDS-polyacrylamide gel (Fig. 2A).
FIGURE 2.
Helicase assays of FANCJ wild-type and mutants on forked duplex and G-quadruplex DNA substrates. Panel A, the purity of the wild-type and mutant FANCJ proteins was evaluated by their detected migration after SDS-PAGE on Coomassie-stained gels according to their predicted sizes. Two micrograms were loaded for each protein. Panel B, helicase reactions (20 μl) were performed by incubating the indicated FANCJ concentration with 0.5 nm forked duplex DNA substrate at 30 °C for 15 min as described under “Experimental Procedures.” Triangle, heat-denatured DNA substrate control. Panel C, helicase reactions were performed as for panel B, except G4 DNA was used instead of forked duplex. M, radiolabeled TP-G4 49-mer oligonucleotide (supplemental Table S1) marker.
First, we examined the helicase activity of these two mutant proteins. Using a 19-bp forked duplex DNA substrate that we used previously (28, 43), FANCJ-WT efficiently unwound it to near completion, whereas R251C and Q255H failed to unwind this type of substrate (Fig. 2B). We also increased the protein amount (96 nm) used in the reaction and the incubation times (1 h), but neither resulted in any detectable helicase activity (data not shown). The inability of the two mutants to unwind the forked duplex substrate was observed with multiple preparations of purified recombinant proteins.
We have found that the FANCJ unwinds 5′ tailed G-quadruplex (G4) DNA substrates (28), and sequential evidence suggested that G4 DNA is a preferred DNA substrate for FANCJ in vitro and in vivo (27, 35, 45, 46). Therefore, we tested the ability of R251C and Q255H to unwind a G4 DNA molecule and found that they failed under conditions where the WT was efficient in this regard (Fig. 2C). In addition, increasing the amount of protein and/or the reaction time did not result in detectable helicase activity (data not shown). Hence, our data indicated that both FANCJ-R251C and -Q255H proteins are unable to unwind a forked duplex or a G4 DNA substrate.
FANCJ-R251C and FANCJ-Q255H Fail to Disrupt Protein-DNA Interactions
Many helicases are not only able to unwind structured DNA but also destabilize DNA-protein complexes. FANCJ has been shown to catalytically disrupt the high affinity interaction of streptavidin bound to a biotinylated ssDNA molecule (47). Therefore, we tested the ability of mutant R215C and Q255H proteins to displace streptavidin bound to a biotinylated oligonucleotide. As shown in Fig. 3, both R251C and Q255H failed to disrupt the streptavidin-biotinylated oligonucleotide interaction under conditions in which WT accomplished >80% streptavidin displacement at a concentration of 19.2 nm. We also increased the amount of protein (96 nm) used in the reaction and the incubation times (1 h), neither of which resulted in any detectable protein displacement activity for the two mutants (data not shown). Thus, we concluded that both FANCJ-R251C and -Q255H lose their ability to disrupt DNA-protein complexes.
FIGURE 3.
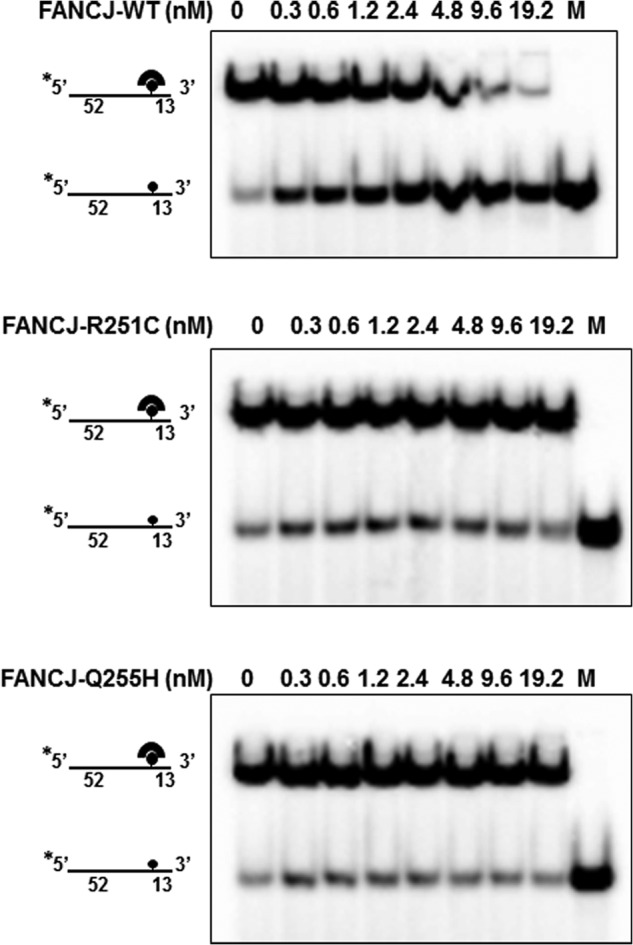
R251C and Q255H mutants are compromised in their ability to displace DNA-bound protein. The indicated concentrations of FANCJ-WT or R251C or Q255H proteins were incubated with 2 mm ATP and DNA substrate (0.5 nm), which had streptavidin bound to the covalently linked biotin moiety residing 52 nucleotides from the 5′end of the radiolabeled oligonucleotide (supplemental Table S1).
FANCJ-Q255H Binds DNA as Wild Type; FANCJ-R251C Binds DNA Poorly
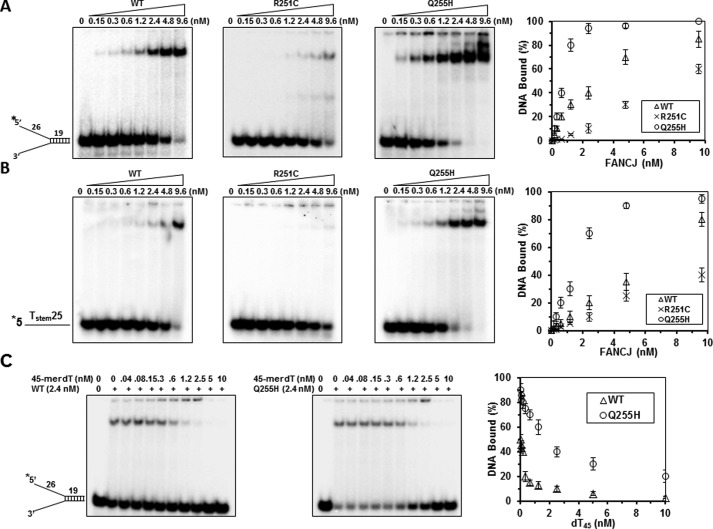
The inability of the two mutants to unwind DNA and disrupt DNA-protein complexes might reflect an impairment of their DNA binding activity. To investigate this possibility, we performed gel mobility shift assays for two mutants and FANCJ-WT with the corresponding radiolabeled forked duplex DNA substrate and an ssDNA oligonucleotide. Qualitative analysis of the DNA binding to either the forked duplex (Fig. 4A) or the single-stranded 44-mer (Tstemstem25, Fig. 4B) demonstrated that the Q255H mutant proteins bound the DNA molecules better than the wild type. For example, 80% of the forked duplex DNA and 70% of the ssDNA bound to Q255H compared with 40 and 20% for the wild-type FANCJ protein at a concentration of 2.4 nm. In contrast, R251C bound both DNA poorly, as only 58% of the forked duplex DNA and 35% of the ssDNA bound R251C protein at the highest concentration (9.6 nm). Q255H also bound the forked duplex or 44-mer ssDNA in the presence of the poorly hydrolysable analog ATPγS or ADP, whereas no stimulation of DNA binding was observed for R251C (data not shown).
FIGURE 4.
DNA binding of mutants and wild-type FANCJ proteins as detected by gel mobility shift assays. Panels A and B, the indicated concentrations of FANCJ-WT, -R251C, or -Q255H protein were incubated with 0.5 nm forked duplex DNA (panel A) or 44-mer ssDNA Tstem25 (panel B) substrate at room temperature for 30 min as described under “Experimental Procedures.” The DNA-protein complexes were resolved on native 5% polyacrylamide gels. Quantitative analyses of DNA gel-shift experiments are shown on the right. Panel C, 2.4 nm FANCJ-WT or -Q255H was incubated with 0.5 nm radiolabeled forked duplex DNA at room temperature for 15 min, and the indicated concentrations of 45-mer ssDNA (dT45) were subsequently added and incubated for an additional 15 min at room temperature. DNA-protein complexes were resolved on native 5% polyacrylamide gels. Quantitative analyses of DNA binding data from DNA competition experiments are shown on the right, with the S.D. indicated by error bars.
To further address the potential DNA binding ability of the Q255H mutant, we performed forked duplex binding experiments in the presence of increasing concentrations of ssDNA competitor molecules. Quantification of the reduction in FANCJ-forked duplex gel-shift species as a function of single-stranded 45-mer competitor (dT45) demonstrated different profiles for the WT and Q255H (Fig. 4C), with Q255H displaying a higher binding stability between the protein and the forked duplex DNA. These results suggested that Q255H binds DNA better than WT.
FANCJ-Q255H Retains ATP Hydrolysis Activity, Whereas FANCJ-R251C Has Poor ATPase Activity
We next examined the DNA-dependent ATPase activity of R251C, Q255H, and WT. Using a covalently closed M13 single-strand DNA as the effector molecule, we determined Km values of ATP hydrolysis for the mutant and wild-type enzymes (Table 1). WT displayed a Km of 0.819 mm, close to the Km value of 0.688 mm determined for Q255H. However, R251C had a higher Km value (4.256 mm), partially due to its poor DNA binding ability. Using an ATP concentration (8.5 mm) ∼10-fold greater than the Km, ATPase assays with the WT and the helicase motif Ia mutants showed that Q255H had a slightly greater kcat for ATP hydrolysis than the WT (17.75 versus 13.31 mm), whereas R251C had a significantly decreased kcat (0.91 mm). We also determined the Keff for M13 ssDNA as a molecular effector; the Keff for WT was 0.087 nm and for Q255H was 0.074 nm, but R251C had a significantly higher Keff (2.942 nm) consistent with its poor DNA binding ability.
TABLE 1.
DNA-dependent ATPase activity of FANCJ-WT and mutants
| Protein | Kma,b | kcata,b,c | kcat/Km | Keff M13 ssDNAa,c |
|---|---|---|---|---|
| mm | s−1 | s−1mm−1 | nm | |
| WT | 0.819 ± 0.092 | 13.31 ± 1.13 | 16.25 | 0.087 ± 0.013 |
| R251C | 4.256 ± 0.749 | 3.88 ± 0.72 | 0.91 | 2.942 ± 0.537 |
| Q255H | 0.688 ± 0.062 | 17.75 ± 1.38 | 25.80 | 0.074 ± 0.018 |
a See “Experimental Procedures” for details.
b M13mp18 ssDNA concentration was 2.1 nm for WT and Q255H and 34 nm for R251C.
c ATP concentration was 8.5 mm.
The Km values suggest that mutant R255H had a similar affinity as the WT for ATP, whereas mutant R251C had a weak interaction with ATP. To directly assess their ability to bind ATP, equal amounts of mutant proteins or WT were incubated with [α-32P]ATP under identical conditions, and binding mixtures were analyzed by size exclusion chromatography. Scintillation counting of the eluted fractions demonstrated that Q255H bound ATP similar to or even more than WT, whereas R251C had less affinity for ATP (Fig. 5). No radiolabeled ATP was retained with BSA, which served as a nonspecific control for ATP binding. The addition of a 10-fold excess of unlabeled ATP outcompeted [32P]ATP binding by either the wild-type or mutant FANCJ proteins (data not shown). Thus, the ATP hydrolysis kinetics assay and physical binding assay suggested that the ATP binding capacity of FANCJ-Q255H is similar to WT, but that of R251C is much lower.
FIGURE 5.
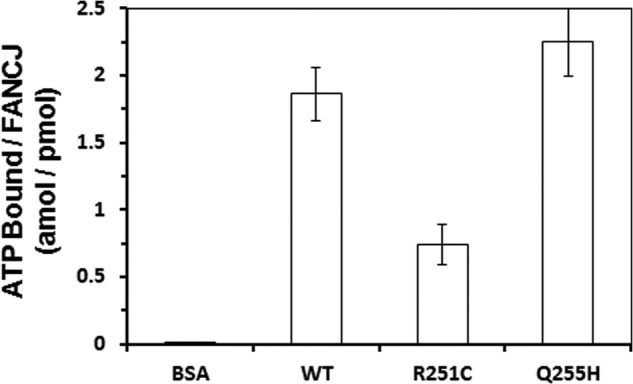
ATP binding by wild-type FANCJ and mutants proteins. [α-32P]ATP binding to FANCJ-WT and mutants was performed by gel filtration chromatography as described under “Experimental Procedures.” The same amount of protein was used, and the total amount of bound ATP was divided by protein and presented as mol ATP/pmol of protein. BSA was used as a control. Statistical analysis of the ATP binding data demonstrated that the difference in p value between FANCJ-WT and -R251C was 0.0007, indicating statistical significance (p < 0.01). The difference in p value between FANCJ-WT and -Q255H was 0.052, indicating that it was not statistically significant (p > 0.05).
FANCJ-Q255H Can Translocate on ssDNA, but FANCJ-R251C Translocates on ssDNA Very Poorly
To examine the effect of mutations on FANCJ translocase activity, we performed two types of experiments with purified FANCJ proteins. We measured DNA-dependent ATPase activity as a function of oligonucleotide (dT) length. To compare the kinetics of ATP hydrolysis between the wild-type and mutant FANCJ proteins, we determined kcat values with increasing lengths of dT oligonucleotides (range 10–200 nucleotides). As expected, kcat values increased with increasing oligonucleotides dT length (Fig. 6A). For FANCJ-Q255H, kcat values increased with increasing oligonucleotide dT length similar to the WT, suggesting that it behaves as an ATP-driven ssDNA translocase. However, FANCJ-R251C displayed a reduced ability to use oligonucleotides as ssDNA effectors for DNA-dependent ATPase activity compared with the WT and Q255H, consistent with its poor ssDNA binding activity.
FIGURE 6.
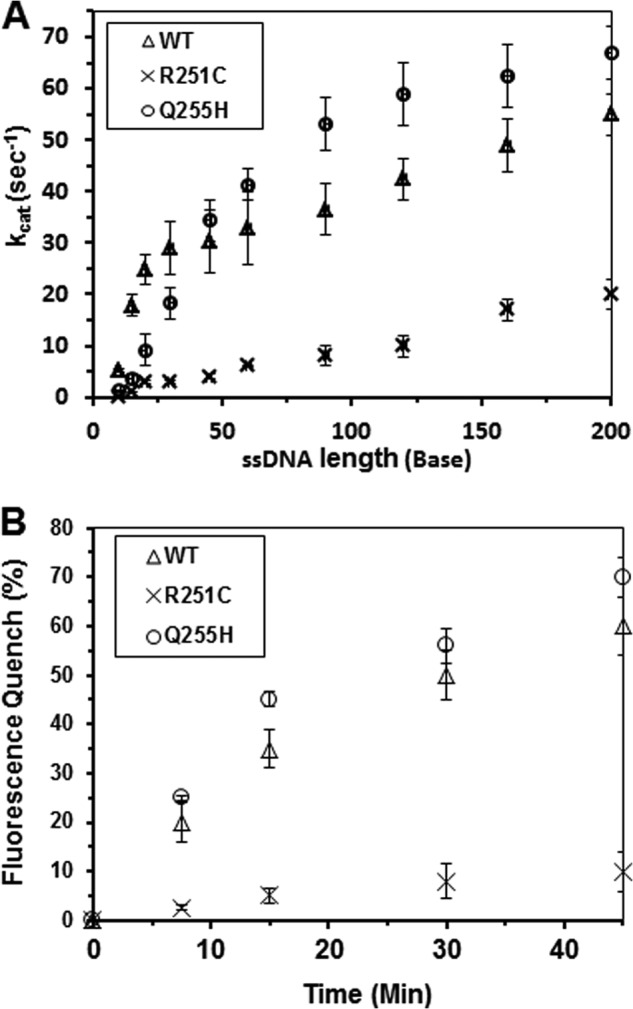
Translocase activity of wild-type FANCJ and mutants proteins. Panel A, the kcat values for ATP hydrolysis catalyzed by FANCJ proteins were determined as a function of oligonucleotide dT length, as described under “Experimental Procedures.” The oligonucleotide dT (length 5–200 nucleotides) concentration ranged from 50 to 5000 nm depending on the length of the oligo dT tested. Error bars represent the S.D. of the fit to the Michaelis-Menten equation. Panel B, fluorescence quenching experiments, performed as described under “Fluorescence Quench Translocation Assays,” using FANCJ-WT and mutants.
To directly test the ability of R251C and Q255H to translocate on ssDNA, we used another approach; that is, a fluorescence quenching assay. Because FANCJ is a 5′ to 3′ helicase, the fluorescence intensity emitted on excitation of the fluorophore covalently bonded to the 3′ end of the DNA molecule will be changed by the close proximity of the helicase to fluorescein. For this purpose we used an identical protein concentration (19.2 nm) and compared fluorescence quench as a function of reaction time with a 3′-fluorescein-conjugated 50-mer oligonucleotide (supplemental Table S1). Our data demonstrated that WT and Q255H had a very similar ability to quench fluorescence in a time-dependent manner (Fig. 6B). However, R251C displayed 6-fold lower translocase activity. In the absence of ATP or in the presence of ATPγS, the increase in fluorescence quench was very small (∼5%) for the longest incubation of WT or Q255H (45 min, data not shown), indicating that ssDNA translocation by either the Q255H mutant or wild-type protein was dependent on hydrolysable ATP.
Both FANCJ-R251C and FANCJ-Q255H Assemble to Dimer as Wild-type FANCJ
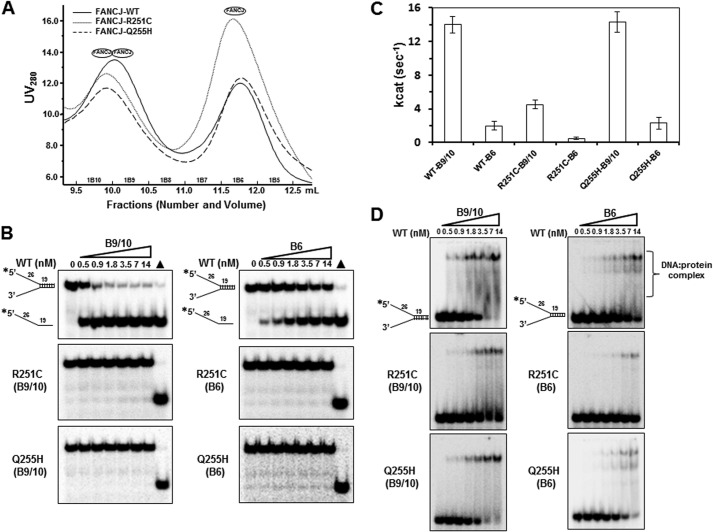
FANCJ is known to form a dimer through its Q motif, and the dimer form has better activity than the monomer form (24). To examine whether R251C and Q255H abolish their helicase activity (both) or DNA binding ability (R251C only) due to their failure to form a dimer, we determined their oligomerization by size exclusion chromatography. Unexpectedly, both R251C and Q255H proteins eluted from a Superdex200 HR column in two major peaks, fractions B9/10 and B6, which represented the dimer and monomer, respectively (Fig. 7A).
FIGURE 7.
Size exclusion chromatography and biochemical analysis of selected FANCJ protein fractions. Panel A, chromatographic profiles of recombinant FANCJ-WT, -R251C, and -Q255H eluting from a Superdex200 HR gel filtration column. Panel B, helicase reactions (20 μl) were performed by incubating the indicated FANCJ fractions with 0.5 nm DNA substrate at 30 °C for 15 min as described under “Experimental Procedures.” Triangle, heat-denatured DNA substrate control. Panel C, ATP hydrolysis (kcat) was determined for each fraction in the presence of covalently closed M13 single-strand DNA (2.1 nm). Statistical analysis of the ATPase data for the B9/10 fraction demonstrated that the difference in p value between FANCJ-WT and -R251C was 0.0001, indicating statistical significance (p < 0.01). The difference in p value between FANCJ-WT and -Q255H was 0.313, indicating that it was not statistically significant (p > 0.05). Similar results were obtained for the three B6 fractions. Panel D, indicated concentrations of FANCJ proteins were incubated with 0.5 nm forked duplex DNA substrate at room temperature for 30 min under standard gel shift assay conditions as described under “Experimental Procedures.” DNA-protein complexes were resolved on native 5% polyacrylamide gels.
The fractions obtained from size exclusion chromatography were further analyzed for helicase activity. Protein titrations demonstrated that the WT B9/10 fraction unwound the 19-bp forked duplex DNA substrate much more efficiently than the WT B6 fraction (Fig. 7B). As expected, both R251C and Q255H fractions lacked any detectable helicase activity.
Furthermore, the six fractions from the three proteins were examined for ATP hydrolysis activity. In the presence of covalently closed M13 ssDNA, the WT B9/10 fraction displayed a 7-fold greater kcat for ATP hydrolysis than the WT B6 fraction (Fig. 7C). The B9/10 and B6 fractions of R251C displayed significantly reduced kcat values (5.0 and 0.5 s−1, respectively) compared with WT B9/10 (15 s−1) and WT B6 (2.2 s−1). However, the two Q255H fractions had similar kcat values to WT.
Moreover, analyses of FANCJ protein binding to forked duplex DNA (Fig. 7D) demonstrated that the WT B9/10 fraction exhibited increased DNA binding compared with the WT B6 fraction throughout the protein titration. For example, WT B9/10 bound 50% of the DNA substrate compared with 15% DNA bound by WT B6 using the same amount of protein (3.5 nm). Although the B9/10 fraction of R251C bound DNA better than the B6 fraction, both poorly bound forked duplex compared with WT. However, both Q255H fractions displayed a similar DNA binding ability to WT.
Both FANCJ-R251C and FANCJ-Q255H Form DNA Damage-induced Foci and Are Recruited to DNA Damage Sites with the Same Kinetics
Failure to restore cisplatin resistance in fancj−/− cells expressing either mutant and the fact that both mutants form dimers similar to wild-type FANCJ led us to explore other possible reasons for these observations in vivo. Therefore, the ability of the mutant proteins to form DNA damage-inducible foci compared with FANCJ-WT was evaluated. We employed the human U2OS cell line for FANCJ localization studies. Cells were transfected with plasmids encoding GFP-FANCJ-WT, GFP-FANCJ-R251C, or GFP-FANCJ-Q255H. Similar to wild-type FANCJ, R251C and Q255H mutants were expressed in the cell nuclei. Cells were then treated with the replication inhibitor hydroxyurea or the DNA cross-linking agent mitomycin C. FANCJ-WT formed nuclear foci after either treatment (Fig. 8A), consistent with previous observations (43). Unexpectedly, both R251C and Q255H mutant proteins localized to nuclei and formed similar foci to FANCJ-WT after treatment with hydroxyurea (HU) or mitomycin C (MMC) (Fig. 8, A and B).
FIGURE 8.
DNA damage response of wild-type and mutants FANCJ proteins in cells. Panel A, GFP fluorescence detection was performed on HeLa cells transfected with plasmids encoding GFP-FANCJ-WT, GFP-FANCJ-R251C, or GFP-FANCJ-Q255H. Cells were treated with 10 mm hydroxyurea (HU) or 1 μm mitomycin C (MMC) for 12 h and fixed for observation under microscope. Panel B, quantitative analyses of GFP-FANCJ foci formation. Data represent the mean of at least 100 cells counted, with the S.D. indicated by error bars. The difference in p value between WT and R251C or Q255H was not statistically significant (p > 0.05) under the three conditions. Panel C, GFP-FANCJ-transfected HeLa cells were treated with a laser (5.5%) at defined regions to induce localized DSBs or incubated with psoralen followed by laser treatment (1.8%) to induce Pso-ICLs. GFP-FANCJ recruitment to Pso-ICLs or DSBs was observed by live-cell imaging using confocal immunofluorescence microscopy 5 min after laser-induced damage.
We next examined whether recruitment of wild-type FANCJ and mutants occurs at the same kinetics under DNA damage circumstances. We used laser-induced DSBs and Pso-ICLs at localized areas within the nuclei of actively dividing transformed HeLa cells. The kinetics of GFP-FANCJ recruitment to the target area was monitored by live cell imaging. We found that R251C and Q255H were recruited to the target area with the same kinetics as WT; the recruitment occurred within 5 min of photoactivation (Fig. 8C) and persisted without apparent diminution for at least 45 min (data not shown). These results suggested that these two FANCJ helicase motif Ia mutants have not impaired their recruitment to DNA damage sites.
Both FANCJ-R251C and FANCJ-Q255H Mutants Exert a Dominant Negative Effect When Expressed in a Wild-type Background
Because both FANCJ-R251C and -Q255H abolished their helicase activity but were recruited to DNA damage sites similar to wild-type protein, we next examined if the FANCJ-R251C or -Q255H mutant allele exerted a dominant negative effect in response to DNA cross-linking agents. Again, we used DT40 cells to express FANCJ proteins. Compared with fancj+/+ cells expressing GFP-tagged FANCJ-WT or transfected with empty vector, fancj+/+ cells expressing FANCJ-R251C or -Q255H showed reduced survival as a function of cisplatin dose and were ∼50-fold more sensitive to the highest concentration of cisplatin (1 μm) (Fig. 9A). γ-H2AX foci were elevated by ∼5-fold in cisplatin-treated fancj+/+ FANCJ-R251C or fancj+/+ FANCJ-Q255H cells compared with the isogenic fancj+/+ FANCJ-WT cells (Fig. 9, B and C). Thus, we concluded that the FANCJ-R251C and -Q255H mutants exert a dominant negative effect on cell survival or the accumulation of DNA damage after cisplatin treatment.
FIGURE 9.
Expression of FANCJ-R251C or FANCJ-Q255H mutant exerts a dominant negative effect on wild-type cells. Panel A, cisplatin sensitivity of cells with indicated genotypes was evaluated by colony formation assay. Panel B, γ-H2AX immunofluorescence staining of cisplatin-treated fancj+/+ cells transfected with plasmids encoding GFP, GFP-FANCJ-WT, GFP-FANCJ-R251C, or GFP-FANCJ-Q255H. DT40 cells were treated with 1 μm cisplatin for 12 h followed by immunofluorescence detection. Panel C, quantitative analyses of γ-H2AX foci shown in Panel B. Data represent the mean of at least 100 cells counted, with the S.D. indicated by error bars. Statistical analysis of the γ-H2AX foci data demonstrated that the differences in p values between fancj+/+ FANCJ-WT cells and fancj+/+ FANCJ-R251C or fancj+/+ FANCJ-Q255H were 0.004 and 0.005, respectively, indicating a significant difference (p < 0.01) for each. Panel D, co-immunoprecipitation experiments using nuclear extracts from HeLa cells expressing GFP, GFP-FANCJ-WT, GFP-FANCJ-R251C, or GFP-FANCJ-Q255H and immunoprecipitated with GFP antibody (GFP-IPed) with the pulldown detected with antibodies against TopBP1, MLH1, and BRCA1, respectively.
FANCJ is known to be more active as a dimer, so one possible mechanism for the dominant negative effect of FANCJ-R251C and -Q255H is that the mutants may function as dominant negatives by dimerizing with the endogenous wild-type enzyme and poisoning its activity. Another possibility is that the mutant protein competes with wild-type FANCJ for interaction with its protein partners (48). Because Western blot analysis demonstrated that GFP-tagged FANCJ-R251C and -Q255H were expressed similar to FANCJ-WT, we wanted to determine whether the FANCJ-R251C and -Q255H variants were associated with proteins known to interact with endogenous wild-type FANCJ. Co-immunoprecipitation experiments using nuclear extracts from HeLa cells demonstrated that TopBP1, MLH1, and BRCA1 proteins previously shown to interact with FANCJ (17, 31, 34) were pulled down by a GFP antibody from lysates of cells that expressed GFP-tagged FANCJ (wild-type and mutants) but not GFP alone (Fig. 9D), suggesting that FANCJ-R251C and -Q255H interact with their protein partners in a manner similar to FANCJ-WT.
DISCUSSION
In many RNA and DNA helicases, motif Ia is believed to play an important role in nucleotide binding; indeed, our biochemical studies of two Fanconi anemia patient mutations both localized in motif Ia indicate that residue Arg-251 is involved in both nucleic acid binding and ATP binding, whereas residue Gln-255 may or may not be implicated in either process (discussed later). Therefore, nucleic acid binding and ATP binding may be executed by a specific residue(s) in FANCJ motif Ia. The reduced ATPase activity of FANCJ-R251C may be partially attributed to its reduced ability to bind DNA (because it is a DNA-stimulated ATPase) and/or bind ATP. Although FANCJ-Q255H retains normal or even enhanced ability to bind DNA and hydrolyze ATPase compared with WT, it is completely inactive as a helicase on short duplex and G-quadruplex DNA substrates. This suggests motif Ia plays an important role in transducing the energy from ATP hydrolysis to the force production required for unwinding structured nucleic acids.
Genetic evidence suggests that some residues, but not every residue, in helicase motif Ia is essential for its biological function. In the yeast DEXD/H-box splicing factor Prp28p, mutation of Arg-264 in motif Ia (262PTRELA267) to Glu or Asp results in lethality (49), suggesting that a positive charge may be important at this position. However, a change of the adjacent residue Glu-265 (from Glu to Gln) can be tolerated to a certain extent. Alanine scanning of motif Ia (146TQPRRVAA153) revealed that only Arg-150 is critical for yeast DEAH-box RNA helicase Prp43 function in vivo (50). In yeast DEAH-box RNA helicase Prp22, however, alanine substitutions of individual amino acids of motif Ia 534TQPRRVAA541 did not impair the biological activity of Prp22; the exception was R538A, which caused a growth defect at 15 °C (51).
Biochemical evidence also demonstrates that some residues in helicase motif Ia are essential, but their exact roles remain debatable. Alanine substitution at Arg-229 in motif Ia (228PRI230) causes severe defects in RNA unwinding that correlate with reduced rates of ATP hydrolysis in the DEAD/DExH helicase NPH-II; however, it retains normal RNA binding ability (52). In Pseudomonas syringae, RecD helicase T259A mutant in motif Ia (258PTGKAAAR265) retains ssDNA binding function. However, the protein displays reduced ATPase activity (72%) and lacks DNA unwinding activity in vitro, but the T259A mutant is able to complement the growth defect of a recD-disrupted strain of P. syringae (53). Retention of the biological activity suggests that the uncoupled ATPase activity in the mutant protein might have other significance. Sulfolobus acidocaldarius XPD motif Ia mutant T56A has reduced ssDNA binding activity and ATP hydrolysis activity and no unwinding activity, suggesting the Thr-56 residue is not critical for DNA binding but is critical for helicase unwinding activity (13). Thus, these residues appear to represent the DNA-binding site that impacts helicase activity by altering XPD binding to DNA or ATP. Alignment of motif Ia in Prp28a, NPH-II, and Prp22 helicases indicates arginine is highly conserved (Fig. 10A), consistent with the critical function of Arg-251 observed in FANCJ helicase.
FIGURE 10.
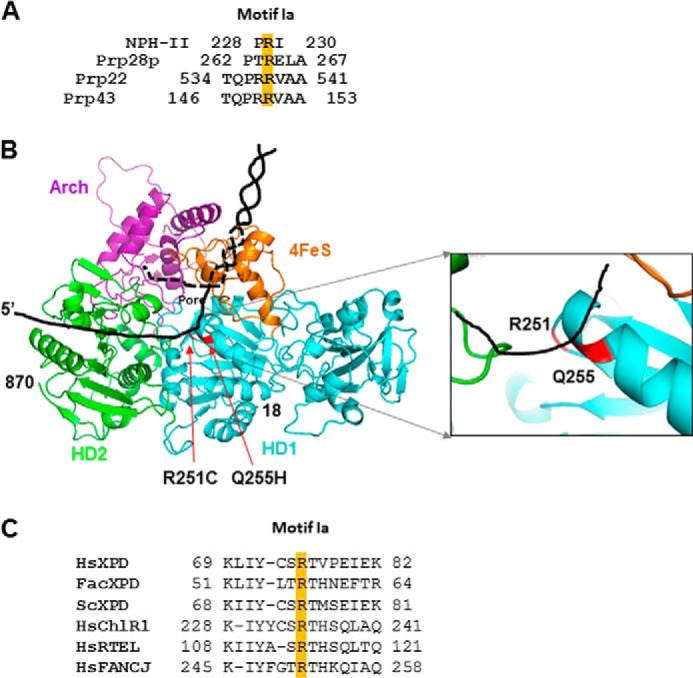
Alignment of helicase motif Ia and model of human FANCJ structure. Panel A, alignment of helicase motif Ia from helicases NPH-II, Prp18p, Prp22, and Prp43. Panel B, mutation sites in the modeled FANCJ structure that are based on SaXPD (23). The helicase domains HD1 (cyan) and HD2 (green), the Fe-S domain (orange), and the Arch domain (purple) are indicated. The position of the pore formed by the four domains is noted. The mutated residues are marked in red and indicated with red arrows. The black line represents DNA, including double helix and ssDNA, and the dashed lines represent DNA behind the protein. Enlargement of the local structure harboring the Arg-251 and Gln-255 residues is shown on the right. Panel C, alignment of helicase motif Ia for FANCJ-sequence like helicases.
Helicase motif Ia has been found to directly contact nucleic acids in the crystal structures of several helicases (8–12, 54). The DNA-protein interactions are predominantly with the phosphodiester backbone of the nucleic acid, as expected for a nonspecific interaction with a single-stranded nucleic acid. In the human FANCJ structure modeled on a S. acidocaldarius XPD crystal structure (13, 23), residues at 251 and 255 sit in the cleft between the two RecA-like domains (Fig. 10B) that have been proposed to function in ATP binding (13). Arg-251 is part of a hairpin loop, and Gln-255 is part of a β sheet (Fig. 10B). Close inspection of the FANCJ helicase family indicates that Arg-251 is in the central position of motif Ia and is highly conserved, whereas Gln-255 is located on its edge and is less conserved (Fig. 10C). Biochemical analyses of purified recombinant FANCJ-Q255H demonstrated that the missense mutation in the motif Ia domain of FANCJ abolished its helicase activity on duplex and G4 DNA substrates, yet the mutant retained its ability to hydrolyze ATP and translocate on ssDNA. Replacement of Arg-251 with a cysteine not only impairs ATP hydrolysis and DNA unwinding activities but also results in DNA binding defects. Because DNA binding is fundamental for FANCJ ATPase and helicase functioning, it is reasonable that the change from a positively charged residue (Arg) to an uncharged medium-sized polar cysteine residue at position 251 and vice versa, from an uncharged glutamine residue to a charged histidine residue at position 255, results in the different and opposing DNA binding abilities observed for FANCJ-R251C and -Q255H proteins. It remains to be determined whether residue Gln-255 is directly involved in DNA and/or ATP binding, although it is likely that Gln-255 contacts DNA and ATP, as suggested by the slightly increased DNA binding and ATP binding activity of Q255H compared with WT (Figs. 4 and 5); however, due to unknown reasons, neither the dimer or monomer fraction of Q255H displays increased binding ability (Fig. 7, C and D). Nevertheless, it will be of interest to know how the Q255H mutant loses its unwinding function. One possible explanation is that the amino acid change results in conformational changes in the two helicase domains (HD1 and HD2) and/or impacts the overall framework stability of FANCJ protein as has been proposed for the structure of SaXPD (13). Therefore, structural determination of human FANCJ with and without DNA will help to determine the exact roles of resides in helicase motif Ia.
Consistent with its effects on the biochemical functions of FANCJ, the FANCJ-R251C and -Q255H mutant alleles failed to rescue cisplatin sensitivity of a fancj null cell line and exerted a dominant negative effect in a wild-type background cell line, as detected by reduced cell survival or induction of γ-H2AX foci after exposure to cisplatin. The basis for the dominant negative phenotypes exerted by the FANCJ-R251C or -Q255H mutant allele is of interest because it may lead to a better understanding of the role of FANCJ in DNA metabolism to preserve genomic stability and enable DNA repair or smooth replication progression to occur. Although it seems likely that FANCJ operates with other proteins of the Fanconi anemia pathway to repair DNA interstrand cross-links, increasing evidence suggests that the helicase performs some of its cellular functions (e.g. resolving G4 structures) outside the classic FA pathway. The dominant negative effect of FANCJ-R251C or -Q255H on cisplatin resistance may be caused by the mutant protein interfering with the proper metabolism of DNA intermediates during replication or repair. Demonstration that both FANCJ-R251C and -Q255H are co-immunoprecipitated with its protein partners BRCA1, MLH1, and TopBP1 suggests that protein hijacking may contribute to the dominant negative phenotypes (48).
Loss of function in a full-length FANCJ protein, such as the helicase defective FANCJ-R251C and -Q255H mutants, may exert biologic effects that are distinguishable from truncated or unstable FANCJ proteins that behave as true nulls. There is a mountain of evidence suggesting that defects in FA pathway genes are linked to an increased risk of cancer (55, 56). Similar to the previously characterized FA patient mutation A349P (43), the R251C and Q255H mutant alleles exerted a dominant negative effect on cell survival or double-strand break formation after cellular exposure to mitomycin C. Formation of FANCJ-R251C and -Q255H foci after cisplatin treatment raises the possibility that the mutant protein disrupts the accumulation or activity of other DNA repair/checkpoint factors at stalled replication forks. The engineered Walker A box K52R mutation that inactivated FANCJ ATPase activity has been shown to exert a dominant negative effect on sensitivity to interstrand cross-linking agents or IR, lead to a delayed entry into the S phase, and activate DNA damage checkpoint machinery (29). However, the K52R site-directed mutation is distinct from the R251C and Q255H patient mutations; the Q255H mutation uncouples DNA translocase activity from helicase activity, whereas R251C has no translocase activity. It will be of interest to determine whether FA carriers that harbor R251C or Q255H allele are characterized by hematopoietic stem cell/progenitor dysfunction or are predisposed to cancers, as such information will be useful to advance diagnosis, prognosis, and treatment of FA and associated breast/ovarian cancer.
Acknowledgment
We thank Dr. Robert Brosh (NIA, National Institutes of Health) for providing various reagents and suggestions.
This work was supported, in whole or in part, by the Saskatchewan Health Research Foundation, the Natural Sciences and Engineering Research Council of Canada, and the Canadian Breast Cancer Foundation (to Y. W.).

This article contains supplemental Table 1.
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- FANCJ
- Fanconi anemia (FA) group J protein
- Pso-ICL
- psoralen-ICL
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- DSB
- double-bond break.
REFERENCES
- 1. Bernstein K. A., Gangloff S., Rothstein R. (2010) The RecQ DNA helicases in DNA repair. Annu. Rev Genet. 44, 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dillingham M. S. (2011) Superfamily I helicases as modular components of DNA-processing machines. Biochem. Soc. Trans. 39, 413–423 [DOI] [PubMed] [Google Scholar]
- 3. Lohman T. M., Bjornson K. P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 4. Lohman T. M., Tomko E. J., Wu C. G. (2008) Non-hexameric DNA helicases and translocases. Mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 9, 391–401 [DOI] [PubMed] [Google Scholar]
- 5. Fairman-Williams M. E., Guenther U. P., Jankowsky E. (2010) SF1 and SF2 helicases. Family matters. Curr. Opin. Struct. Biol. 20, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pyle A. M. (2008) Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 7. Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 8. Korolev S., Hsieh J., Gauss G. H., Lohman T. M., Waksman G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90, 635–647 [DOI] [PubMed] [Google Scholar]
- 9. Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., Wigley D. B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 [DOI] [PubMed] [Google Scholar]
- 10. Kim J. L., Morgenstern K. A., Griffith J. P., Dwyer M. D., Thomson J. A., Murcko M. A., Lin C., Caron P. R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide. The crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 [DOI] [PubMed] [Google Scholar]
- 11. Yao N., Hesson T., Cable M., Hong Z., Kwong A. D., Le H. V., Weber P. C. (1997) Structure of the hepatitis C virus RNA helicase domain. Nat. Struct Biol. 4, 463–467 [DOI] [PubMed] [Google Scholar]
- 12. Büttner K., Nehring S., Hopfner K. P. (2007) Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct Mol Biol. 14, 647–652 [DOI] [PubMed] [Google Scholar]
- 13. Fan L., Fuss J. O., Cheng Q. J., Arvai A. S., Hammel M., Roberts V. A., Cooper P. K., Tainer J. A. (2008) XPD helicase structures and activities. Insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H., Rudolf J., Johnson K. A., McMahon S. A., Oke M., Carter L., McRobbie A. M., Brown S. E., Naismith J. H., White M. F. (2008) Structure of the DNA repair helicase XPD. Cell 133, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolski S. C., Kuper J., Hänzelmann P., Truglio J. J., Croteau D. L., Van Houten B., Kisker C. (2008) Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 6, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marintcheva B., Weller S. K. (2003) Helicase motif Ia is involved in single-strand DNA-binding and helicase activities of the herpes simplex virus type 1 origin-binding protein, UL9. J. Virol. 77, 2477–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160 [DOI] [PubMed] [Google Scholar]
- 18. Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K., North B., McGuffog L., Evans D. G., Eccles D., Breast Cancer Susceptibility Collaboration (UK), Easton D. F., Stratton M. R., Rahman N. (2006) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 38, 1239–1241 [DOI] [PubMed] [Google Scholar]
- 19. Levitus M., Waisfisz Q., Godthelp B. C., de Vries Y., Hussain S., Wiegant W. W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M. A., Pals G., Arwert F., Mathew C. G., Zdzienicka M. Z., Hiom K., De Winter J. P., Joenje H. (2005) The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37, 934–935 [DOI] [PubMed] [Google Scholar]
- 20. Levran O., Attwooll C., Henry R. T., Milton K. L., Neveling K., Rio P., Batish S. D., Kalb R., Velleuer E., Barral S., Ott J., Petrini J., Schindler D., Hanenberg H., Auerbach A. D. (2005) The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37, 931–933 [DOI] [PubMed] [Google Scholar]
- 21. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. (2005) BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8, 255–265 [DOI] [PubMed] [Google Scholar]
- 22. Rafnar T., Gudbjartsson D. F., Sulem P., Jonasdottir A., Sigurdsson A., Jonasdottir A., Besenbacher S., Lundin P., Stacey S. N., Gudmundsson J., Magnusson O. T., le Roux L., Orlygsdottir G., Helgadottir H. T., Johannsdottir H., Gylfason A., Tryggvadottir L., Jonasson J. G., de Juan A., Ortega E., Ramon-Cajal J. M., García-Prats M. D., Mayordomo C., Panadero A., Rivera F., Aben K. K., van Altena A. M., Massuger L. F., Aavikko M., Kujala P. M., Staff S., Aaltonen L. A., Olafsdottir K., Bjornsson J., Kong A., Salvarsdottir A., Saemundsson H., Olafsson K., Benediktsdottir K. R., Gulcher J., Masson G., Kiemeney L. A., Mayordomo J. I., Thorsteinsdottir U., Stefansson K. (2011) Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 43, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 23. Wu Y., Suhasini A. N., Brosh R. M., Jr. (2009) Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell. Mol. Life Sci. 66, 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Y., Sommers J. A., Loiland J. A., Kitao H., Kuper J., Kisker C., Brosh R. M. (2012) The Q motif of FANCJ DNA helicase regulates its dimerization, DNA binding, and DNA repair function. J. Biol. Chem. 287, 21699–21716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D. M. (2004) The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. U.S.A. 101, 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta R., Sharma S., Sommers J. A., Jin Z., Cantor S. B., Brosh R. M., Jr. (2005) Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 280, 25450–25460 [DOI] [PubMed] [Google Scholar]
- 27. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., Hiom K. (2008) FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283, 36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Y., Shin-ya K., Brosh R. M., Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumaraswamy E., Shiekhattar R. (2007) Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 27, 6733–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R., Sharma S., Sommers J. A., Kenny M. K., Cantor S. B., Brosh R. M., Jr. (2007) FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 110, 2390–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peng M., Litman R., Xie J., Sharma S., Brosh R. M., Jr., Cantor S. B. (2007) The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 26, 3238–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Suhasini A. N., Rawtani N. A., Wu Y., Sommers J. A., Sharma S., Mosedale G., North P. S., Cantor S. B., Hickson I. D., Brosh R. M., Jr. (2011) Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 30, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suhasini A. N., Sommers J. A., Muniandy P. A., Coulombe Y., Cantor S. B., Masson J. Y., Seidman M. M., Brosh R. M., Jr. (2013) Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell. Biol. 33, 2212–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gong Z., Kim J. E., Leung C. C., Glover J. N., Chen J. (2010) BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell 37, 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwab R. A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. (2013) FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 201, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y., Brosh R. M., Jr. (2009) FANCJ helicase operates in the Fanconi anemia DNA repair pathway and the response to replicational stress. Curr. Mol. Med. 9, 470–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frank B., Hemminki K., Meindl A., Wappenschmidt B., Sutter C., Kiechle M., Bugert P., Schmutzler R. K., Bartram C. R., Burwinkel B. (2007) BRIP1 (BACH1) variants and familial breast cancer risk. A case-control study. BMC. Cancer 7, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis A. G., Flanagan J., Marsh A., Pupo G. M., Mann G., Spurdle A. B., Lindeman G. J., Visvader J. E., Brown M. A., Chenevix-Trench G. (2005) Mutation analysis of FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast Cancer Res. 7, R1005–R1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rutter J. L., Smith A. M., Dávila M. R., Sigurdson A. J., Giusti R. M., Pineda M. A., Doody M. M., Tucker M. A., Greene M. H., Zhang J., Struewing J. P. (2003) Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from breast-ovarian cancer families and among early-onset breast cancer cases and reference individuals. Hum. Mutat. 22, 121–128 [DOI] [PubMed] [Google Scholar]
- 40. Sigurdson A. J., Hauptmann M., Chatterjee N., Alexander B. H., Doody M. M., Rutter J. L., Struewing J. P. (2004) Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vahteristo P., Yliannala K., Tamminen A., Eerola H., Blomqvist C., Nevanlinna H. (2006) BACH1 Ser919Pro variant and breast cancer risk. BMC Cancer 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cao A. Y., Huang J., Hu Z., Li W. F., Ma Z. L., Tang L. L., Zhang B., Su F. X., Zhou J., Di G. H., Shen K. W., Wu J., Lu J. S., Luo J. M., Yuan W. T., Shen Z. Z., Huang W., Shao Z. M. (2009) Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res. Treat. 115, 51–55 [DOI] [PubMed] [Google Scholar]
- 43. Wu Y., Sommers J. A., Suhasini A. N., Leonard T., Deakyne J. S., Mazin A. V., Shin-Ya K., Kitao H., Brosh R. M., Jr. (2010) Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood 116, 3780–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bridge W. L., Vandenberg C. J., Franklin R. J., Hiom K. (2005) The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37, 953–957 [DOI] [PubMed] [Google Scholar]
- 45. Sarkies P., Murat P., Phillips L. G., Patel K. J., Balasubramanian S., Sale J. E. (2012) FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 40, 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Henderson A., Wu Y., Huang Y. C., Chavez E. A., Platt J., Johnson F. B., Brosh R. M., Jr., Sen D., Lansdorp P. M. (2014) Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 42, 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sommers J. A., Rawtani N., Gupta R., Bugreev D. V., Mazin A. V., Cantor S. B., Brosh R. M., Jr. (2009) FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 284, 7505–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Y., Brosh R. M., Jr. (2010) Helicase-inactivating mutations as a basis for dominant negative phenotypes. Cell Cycle 9, 4080–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang T. H., Latus L. J., Liu Z., Abbott J. M. (1997) Genetic interactions of conserved Rrgions in the DEAD-Box protein Prp28p. Nucleic Acids Res. 25, 5033–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tanaka N., Schwer B. (2006) Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45, 6510–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneider S., Campodonico E., Schwer B. (2004) Motifs IV and V in the DEAH Box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 279, 8617–8626 [DOI] [PubMed] [Google Scholar]
- 52. Gross C. H., Shuman S. (1998) The nucleoside triphosphatase and helicase activities of vaccinia virus NPH-II are essential for virus replication. J. Virol. 72, 4729–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Satapathy A. K., Pavankumar T. L., Bhattacharjya S., Sankaranarayanan R., Ray M. K. (2008) ATPase activity of RecD is essential for growth of the Antarctic Pseudomonas syringae Lz4W at low temperature. FEBS J 275, 1835–1851 [DOI] [PubMed] [Google Scholar]
- 54. Kuper J., Wolski S. C., Michels G., Kisker C. (2012) Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 31, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kottemann M. C., Smogorzewska A. (2013) Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deans A. J., West S. C. (2011) DNA interstrand crosslink repair and cancer. Nat. Rev Cancer 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]