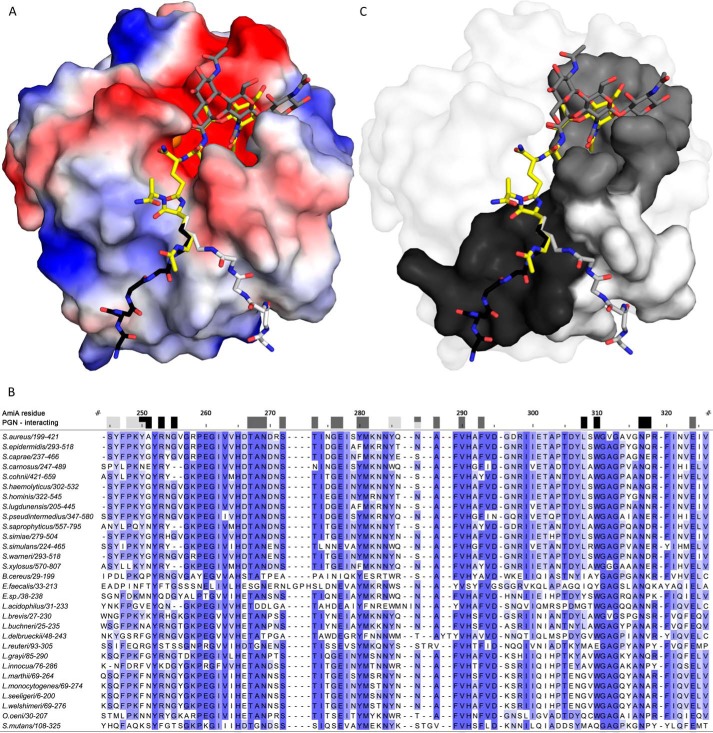
FIGURE 5.
AmiA-cat binding to PGN. A, electrostatic surface of AmiA-cat with MtetP (yellow), zinc (orange), and modeled PGN components (dark gray, black, and light gray). Uncharged (white surface area), positively charged (blue areas), and negatively charged residues (red surface) are shown. The spacious hydrophilic pocket harboring the zinc ion and active site also accommodates MurNAc. Adjacent GlcNAc rings (dark gray sticks) shift MurNAc slightly when modeled as a polymer. The lower peptide moiety of MtetP binds in the mostly uncharged region of the binding cleft. Two conformations for the pentaglycine bridge (black and light gray sticks, respectively) linked to l-Lys of MtetP were modeled according to uncharged surface area and possible hydrogen bonds. B, a multisequence alignment of bacterial amidases with identical residues colored by conservation from light to dark blue. Residues forming the carbohydrate binding pocket, marked by dark gray boxes, are highly conserved among all compared amidases. Surface-exposed amino acids near the two pentaglycine bridges are marked in black and light gray for the respective conformation models. Conservation, especially among staphylococci is high for both. Alignment was calculated using Clustal Omega (58), and the output was created using Jalview (59). C, conserved residues mapped on the AmiA-cat surface according to the color scheme used in A and B.