FIGURE 8.
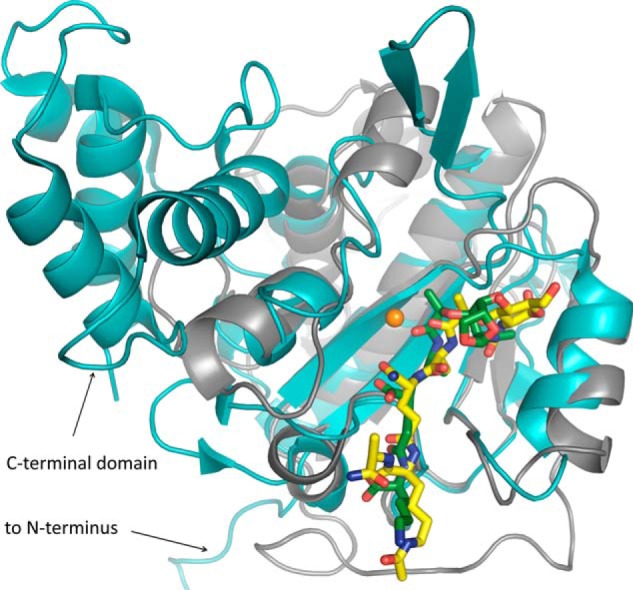
Comparison of the AmiA-cat complex (gray schematic) with AmiD from E. coli (cyan schematic; Protein Data Bank code 3D2Y). Superimposition of the two bacterial amidases reveals a similar fold solely around the binding and active site. AmiD deviates by a 2.0-Å root mean square deviation (DaliLite pairwise) from the AmiA-cat main chain, amino acids in the active and binding sites differ, and AmiD contains additional motifs at its N and C termini. Ligand positioning of anhydro-MTP (green sticks) to AmiD is comparable with the AmiA-cat complex with MtetP (yellow sticks). However, anhydro-MTP has an overall shift in relation to MtetP, and interactions of enzyme with ligand are unalike. Additionally, the MurNAc moiety, including the scissile bond, lies in the direct vicinity of the zinc binding residues and is in the anhydro form, which does not occur in staphylococci. Zn2+ from the unliganded AmiA-cat structure in orange was superimposed.
