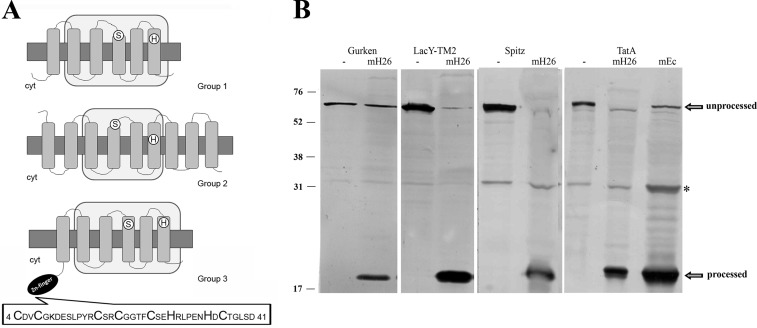
FIGURE 1.
Haloarchaeal Rho display conserved active site residues and membrane associated Rho activity. A, the translated amino acid sequences of Rho homologs were introduced in the TMHMM and SMART programs, and the retrieved topologies and domain architectures are schematically represented. Top, E. coli GlpG protein, representing Group 1. Middle, Hvo_1474, (Group 2). Bottom, Hvo_0727 (Group 3). Gray rectangles, TMDs; light gray rectangles, Rho domains; white circles, positions of active site serine and histidine; black ellipse, cytoplasmic zinc finger motif. The sequence of the zinc finger motif is shown at the bottom. Cyt, cytoplasm. B, cleavage of specific Rho substrates by H. volcanii solubilized membrane fractions. Cell extracts of E. coli MG1655 transformed with plasmid pKS505 (Gurken), pKS506 (LacY-TMD2), pKS507 (Spitz), or pKS508 (TatA) were incubated with (mH26) or without (−) solubilized membranes of H. volcanii H26. As a cleavage site control, cell extract of pKS508 incubated with E. coli Rossetta solubilized membranes (mEc) is shown on the right. After incubation, samples were TCA-precipitated and used in Western blotting experiments with anti-His antibody. The bands corresponding to unprocessed and processed substrates are pointed out by arrows. *, a nonspecific protein from E. coli extracts that cross-reacts with the anti-His antibody. Migration of molecular mass markers (kDa) is indicated at the left.