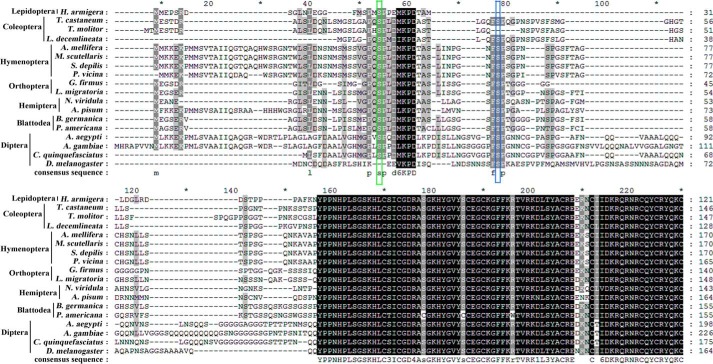
FIGURE 9.
Alignment of H. armigera USP1 with USP1 from other insects. USP1 amino acids sequences were performed alignment from different insects, including H. armigera (ACD74808.1), Tribolium castaneum (NP_001107766.2), Tenebrio molitor (CAB75361.1), Leptinotarsa decemlineata (BAD99298.1), Apis mellifera (NP_001011634.1), Melipona scutellaris (AAW02952.1), Scaptotrigona depilis (ABB00308.1), Polyrhachis vicina (AGF50212.1), Gryllus firmus (ADL09403.1), Locusta migratoria (AAQ55293.1), Nizara viridula (ADQ43369.1), Acyrthosiphon pisum (NP_001155140.1), Blattella germanica (CAH69897.1), Periplaneta americana (BAM63276.2M.), Aedes aegypti (AAG24886.1), Anopheles gambiae (XP_320944.5), Culex quinquefasciatus (XP_001866328.1), D. melanogaster (AAF45707.1). The green box indicates the phosphorylation site identified in this study in H. armigera USP1 at Ser-21, and the blue box indicates the USP phosphorylation site Ser-35 in D. melanogaster. These data just show the alignment of N-terminal amino acids.