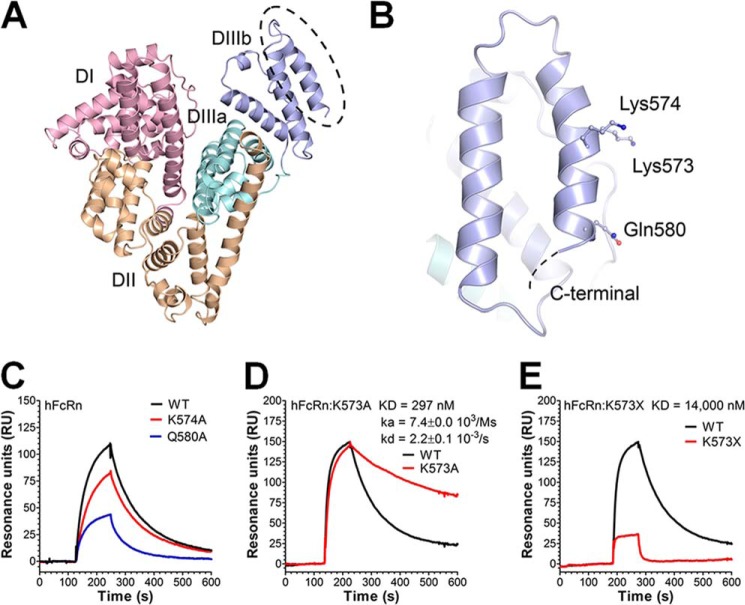
FIGURE 1.
Amino acid substitutions in the last C-terminal end of HSA DIII modulate binding to hFcRn. A, an illustration of the crystal structure of full-length HSA with the three domains DI (pink), DII (orange), and DIII (cyan/blue) highlighted. The DIII is split into subdomains DIIIa (cyan) and DIIIb (blue). B, a close-up of DIIIb and the last α-helix in the C-terminal end with the amino acid residues Lys-573, Lys-574, and Gln-580 indicated. The figures were made using the software PyMOL, using coordinates from Protein Data Bank entry 1bm0. C–E, representative SPR sensorgrams showing binding of 1 μm of WT HSA, K574A, and Q580A (C); WT HSA and K573A (D); and WT HSA and HSA K573X (E) to immobilized hFcRn at pH 6.0. Injections were performed at 25 °C, and the flow rate was set to 40 μl/min. The kinetic rate constants were obtained using a simple first order (1:1) bimolecular interaction model (Langmuir) or a steady state affinity model supplied by the BIAevaluation 4.1 software. The kinetic values represent the average of triplicates.