Figure 6.
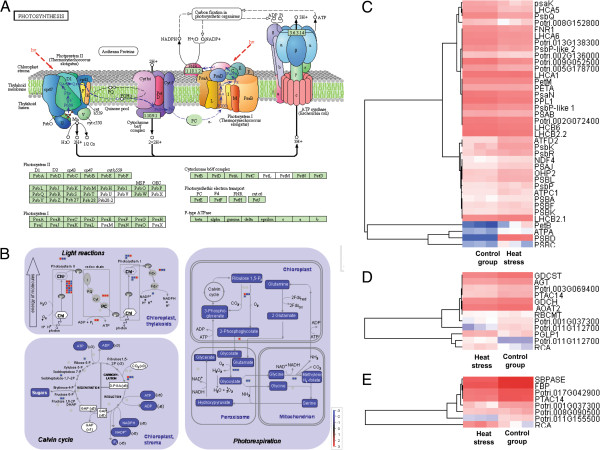
Diagram of differentially expressed genes involved in photosynthesis. A: photosynthesis pathway (reference KEGG) B: The ‘photosynthesis’ MapMan pathway was used to visualize transcriptional changes in genes with putative functions in metabolism. Red represents higher expression in heat stress samples and blue denotes higher expression in controls, with darker shading indicating increasing magnitude of log2 expression fold change, as specified by the scale. C: Pearson correlation coefficient heat map indicating the differentially expressed genes related to photosynthesis. D: Pearson correlation coefficient heat map indicating the differentially expressed genes related to photorespiration. E: Pearson correlation coefficient heat map indicating the differentially expressed genes related to calvin cycle. Red and blue indicate higher and lower transcript levels, respectively. The gene model is shown on the right. The control group consisted of three biological samples that were not treated with high temperature.
