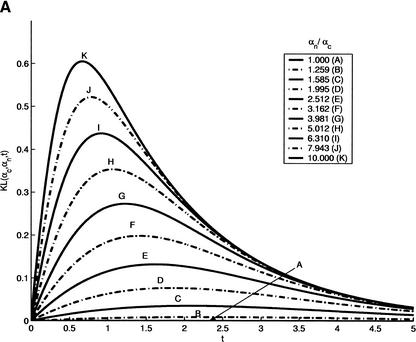
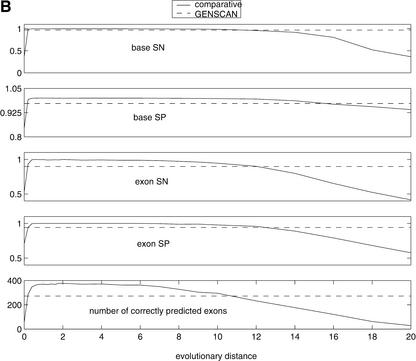
Figure 9.
(A) Precision (KL-divergence in the figure) as a function of divergence time. Through evolution, coding and noncoding regions are characterized with Jukes-Cantor substitution models with parameters αc and αn, respectively. We set αc = 0.1and the ratio of αn/αc is sampled from 1–10. Larger differences between parameters imply shorter optimal time t* required for minimal prediction error. Similarly, the more different the conservation rates of the two regions are, the higher the precision is. (B) Performance of human gene prediction was evaluated on synthetic orthologs at different evolutionary distances. The evolutionary distance of 1 corresponds to the distance between human and mouse. In all cases, comparative gene finder outperforms GENSCAN (indicated by broken lines) on the plateau between the distances of 0.5 and 6. Performance degrades significantly for very low and very high distances.