Figure 3.
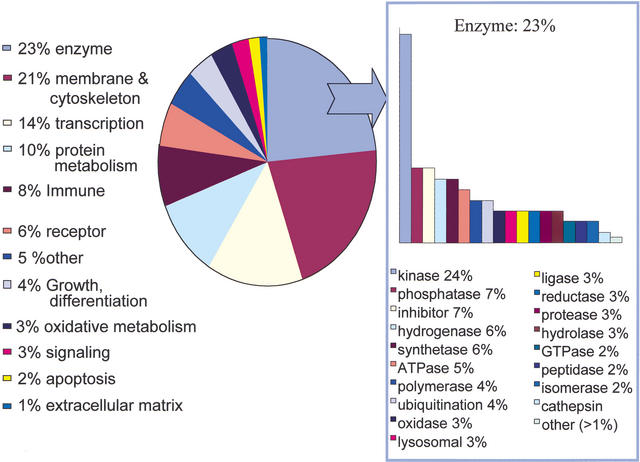
Functional analysis of known transcripts in the immunome libraries; 4284 known gene clusters were represented and analyzed using FANTOM2 GO terms extracted from the TC descriptor. Enzymes were the largest category identified, with 1018 unique members. The cytoskeleton category contained 916 members and included membrane bound proteins (excluding receptors), the actin cytoskeleton, and trafficking molecules. Three hundred ninety transcriptional regulators were identified. Three hundred forty-nine TC annotated as immunity/antigen presentation were specifically involved in MHC processing, antigen presentation, chemokine, leukotriene and cytokine production, components of the complement cascade, and interferon signaling. One hundred seventy-two cell cycle, growth and differentiation TC included a number of macrophage-specific, neutrophil-specific, and T-cell-specific differentiation markers. G-proteins, kinases, and the rab/rac signaling pathways were particularly represented in the 108 signaling TCs. The remaining categories included protein synthesis and modification (433 TC), 243 receptors, oxidative metabolism/stress (139 TC), apoptosis (68 TC), and extracellular matrix (52TC).