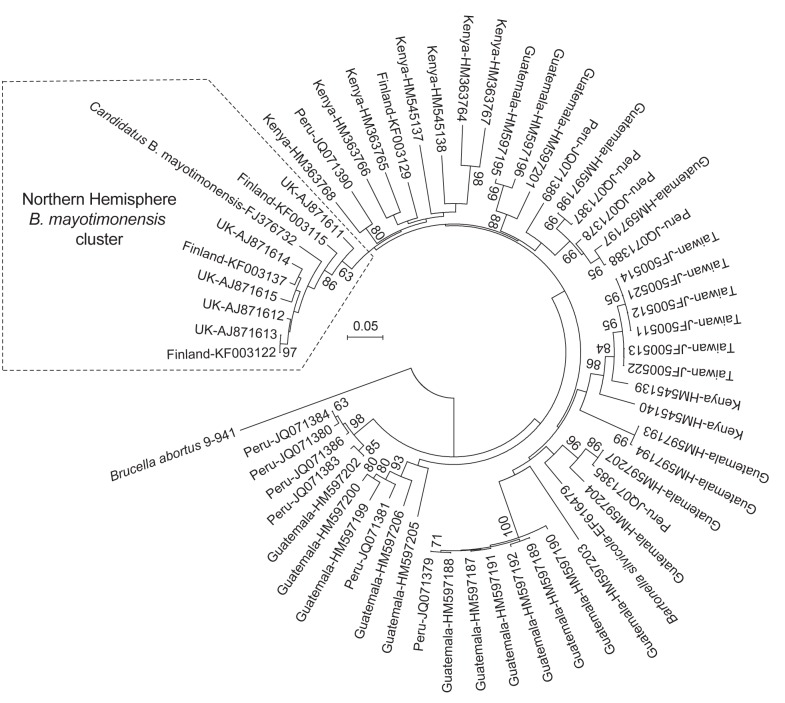
Figure 3.
Phylogenetic analysis of bat-colonizing Bartonella spp. found worldwide demonstrates a distinct B. mayotimonensis cluster in the Northern Hemisphere. Maximum composite likelihood–based neighbor-joining tree is based on the alignment of the gltA multilocus sequence analysis marker. Information from Bartonella gltA sequences from bat blood isolates or from minced tissues of bats or from bat ectoparasites was included in the analysis. GenBank accession numbers of the sequences are shown after the country of origin. Numbers on branches indicate bootstrap support values derived from 1,000 tree replicas. Bootstrap values >60 are shown. Scale bar indicates nucleotide substitutions per site.