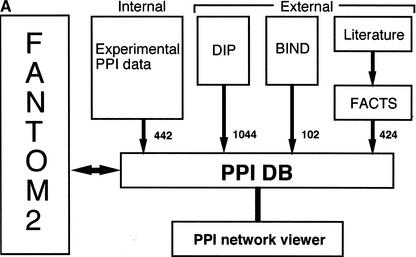
Figure 1.
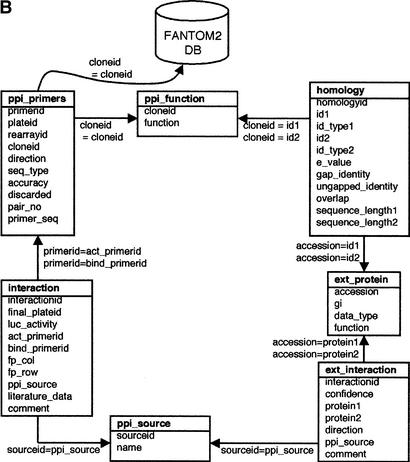
RIKEN PPI database. (A) Overview of the database. The number of interactions obtained from each data source is shown near the arrows. The interactions from DIP and BIND public database are updated regularly, and the interactions from the internal experiments and FACTS are also updated in a timely fashion. (B) The table structure of the PPI database. Each box represents a table and contains the table name at the top, followed by the column names. The arrows and equation labels indicate corresponding column names between two tables. Using this correspondence, exactly one entry in the table at the end of the arrow can be identified by using a corresponding value in the column of the table at the beginning of the arrow. A specific entry in the FANTOM2 database can be retrieved by using the clone ID as a key. The various tables are:
- ppi_primers: Contains primer IDs as well as primer sequences and clone IDs. A protein sequence can be estimated by using this table and the RIKEN cDNA database.
- ppi_function: Includes the functional annotation for each RIKEN clone.
- interaction: Comprises a list of interactions identified through our assays (internal PPIs).
- ext_interaction: Stores a list of interactions extracted from publicly available data sources (external PPIs).
- ext_protein: Includes protein information of the external PPIs. Each protein is identified by its NCBI accession number.
- ppi_source: Stores information regarding the origin of the interactions, i.e., RIKEN_PPI assay, DIP, BIND, or literature.
- homology: Contains the results of homology searches and the alignment of two sequences.