Figure 2.
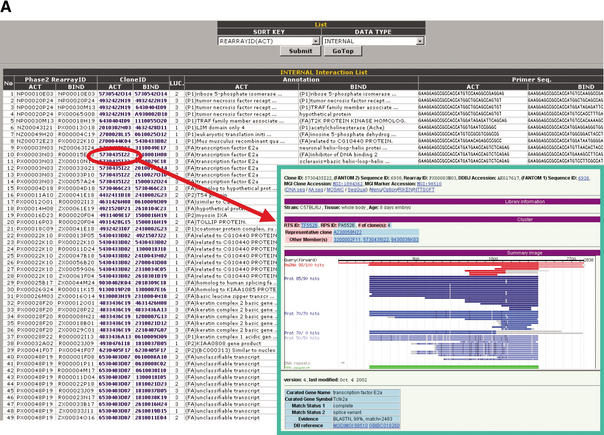
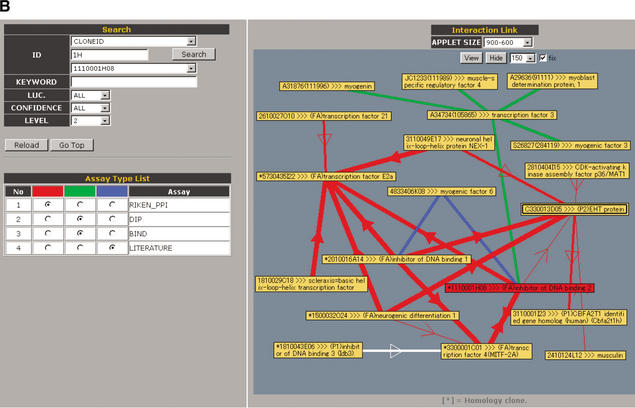
Screen display of the PPI Viewer. The viewer consists of the Interaction List (A) and the Interaction Network (B). The Interaction List displays all the deposited PPIs sorted according to the proteins' IDs. For internal PPIs, the list can be sorted according to bait (BIND) or prey (ACT) proteins. The listed data include the interaction pairs and their annotations, supplemented with information of the primer sequences for construction of the assay samples and the luciferase activity (LUC) of the interaction (internal PPIs) or the data source linked to the MEDLINE abstracts (external PPIs). Clone IDs in the list are linked to the main FANTOM2 viewer (right lower inset). Some clone IDs are linked to the RIKEN 3′-end-sequence database with homology information because we have used several RIKEN clones that correspond to mouse known genes and therefore have not been fully sequenced. Accession IDs (GI number) in the external interaction list are linked to the NCBI clone viewer. These links enable users to obtain detailed information on the clone. The Interaction Network (B) comprises three sections labeled Search, Assay Type List, and Interaction Link. In the Search section, users can search for a clone to display according to any part of letters or numbers in its clone ID, accession ID, or GI number. Users also can search for proteins by KEYWORD. For example, entering “death” as a keyword yields clone 2010323J02 (programmed cell death 6). LUC, CONFIDENCE, and LEVEL are parameters with which users can display interactions having the desired luciferase activity, confidence and within the desired number of interaction steps from the target protein, respectively. In the Assay Type List section, interactions from selected data sources can be color-coded (red, green, or blue). In the Interaction Link section, the protein interaction network for the target protein (shown as a red node) is displayed as edges and nodes. The luciferase activity (LUC) and confidence of an interaction are reflected by the thickness of its edge. For internal PPIs, an arrow indicates the direction of bait protein to prey protein in the mammalian two-hybrid assay. Self-interactions are shown as double boxes, and integrated genes are shown as a single node with an asterisk. The interaction network is labeled with IDs (default) or with both IDs and annotations (by clicking the View and Hide buttons). The APPLET SIZE and length of edges (pulldowns to the right of the Hide button) can be changed. Brief instructions are listed in Table 1.