Figure 4.
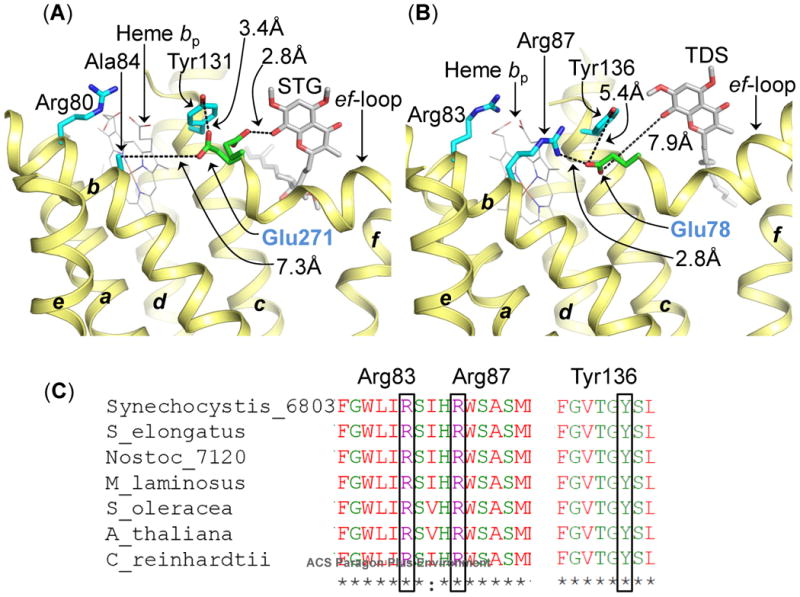
Role of conserved Glu (Pro-Glu-Trp-Tyr) in semiquinone deprotonation in cyochrome bc complexes. (A) In the bc1 complex, the Glu271 residue (green/red stcks, labeled in blue) of the PEWY sequence is found to occupy two distinct locations-quinone proximal (PDB ID 3CX5) and heme bp-proximal (PDB ID 1NTZ). In the heme bp-proximal position, Glu271 inteacts with Tyr131 (cytochrome b polypeptide). The figure was generated by superposition of cytochrome bc1 crystal structures (PDB IDs 1NTZ and 3CX5). The STG molecule and the quinone/STG-proximal Glu271 orientation were obtained from PDB ID 3CX5. (B) In the cytochrome b6f complex crystal structure (PDB ID 4H44), Glu78 (green/red sticks, labeled in blue) of subunitIV, homologous to Glu271 in the bc1 complex, is located in the heme bp-proximal orientation. In this position, Glu78 interacts with Arg87 of cytochrome b6 (b6f), through a distance of 2.8 Å. Arg87 (b6f) is replaced by Ala84 (cyan sticks) in the bc1 complex. Arg83 of cytochrome b6 (b6f) and Arg80 of cytochrome b (bc1) are conserved in their location. The quinone analog inhibitor TDS has been inserted into the figure from PDB ID 4H13 to mark the Qp-site. The transmembrane helices (a-g) are labeled. The polypeptides are shown as ribbons. (C) Multiple sequence alignment of the cytochrome b6 subunit of the cytochrome b6f complex from prokaryotic cyanobacteria (Synechocystis PCC 6803, Synechococcus elongates PCC 6301, Nostoc PCC 7120, M. laminosus), a eukaryotic alga (C. reinhardtii), and higher plants (Arabidopsis thaliana and Spinacea oleracea). Arg83, Arg87 (trans-membrane helix B) and Tyr136 (trans-membrane helix C) are conserved in cytochrome b6 polypeptide.
