Abstract
We performed genome-wide sequence comparisons at the protein coding level between the genome sequences of Drosophila melanogaster and Anopheles gambiae. Such comparisons detect evolutionarily conserved regions (ecores) that can be used for a qualitative and quantitative evaluation of the available annotations of both genomes. They also provide novel candidate features for annotation. The percentage of ecores mapping outside annotations in the A. gambiae genome is about fourfold higher than in D. melanogaster. The A. gambiae genome assembly also contains a high proportion of duplicated ecores, possibly resulting from artefactual sequence duplications in the genome assembly. The occurrence of 4063 ecores in the D. melanogaster genome outside annotations suggests that some genes are not yet or only partially annotated. The present work illustrates the power of comparative genomics approaches towards an exhaustive and accurate establishment of gene models and gene catalogues in insect genomes.
Whole-genome sequence comparisons between genomes from metazoans can be used to detect sequence conservation both in coding and noncoding regions. Whereas conservation of coding regions can be detected between species separated by large evolutionary distances (e.g., between mammals and fish; Roest Crollius et al. 2000), the conservation of noncoding regions is usually much weaker and mainly detected between species that are separated by shorter evolutionary distances (e.g., within mammals; Kent 2002; Mural et al. 2002). In other words, the kind and amount of information that can be deduced from genomic DNA comparisons depend on the evolutionary distance between the species.
The annotation process used for Drosophila (Rubin et al. 2000) relied on protein database searches, cDNA, and EST matches and ab initio gene predictions. The power of protein comparisons was high, but not exhaustive, because they concerned mainly species such as yeast, Caenorhabditis elegans and mammals that are relatively distant from the fruit fly. However, ab initio predictions and cDNA sequencing could notably complement the annotations beyond conserved genes, and a total of 13,666 genes was proposed for the analysis of the fly genome (Adams et al. 2000; Misra et al. 2002). While finishing and analysis of the fly genome sequence was still in progress, an additional set of genes was proposed (Gopal et al. 2001). The establishment of a draft sequence of the genome of Anopheles gambiae (Holt et al. 2002) offers the possibility of reevaluation of the present D. melanogaster gene inventory using a rationale that we used previously to compare a fraction of the human genome to that of a teleost fish, Tetraodon nigroviridis (Roest Crollius et al. 2000). Conversely, it will also provide an evaluation of the initial Anopheles genome annotations. We therefore carried out this type of global comparison between these two insect genomes.
RESULTS AND DISCUSSION
The Drosophila Annotation
The Exofish procedure (for EXOn FInding by Sequence Homology) that we developed for large-scale genome comparisons is based on the BLAST algorithm (Altschul et al. 1990). To minimize background of false positive alignments outside coding regions and to maximize the detection of evolutionarily conserved regions (ecores), TBLASTX parameters and filter conditions were adjusted on a set of reference sequences (see Methods).
The available sequence assembly of A. gambiae (http://www.ensembl.org/Anopheles_gambiae) and the last two versions of the D. melanogaster genome (http://www.fruitfly.org/annot/release2.html and http://www.fruitfly.org/annot/release3.html) were compared using the adjusted settings of Exofish. A whole-genome comparison between the two genomes resulted in a total of 47,134 ecores (for release 2) or 46,742 ecores (for release 3) in the D. melanogaster genome (Table 1; available at www.genoscope.cns.fr/Exofish/Fly). These numbers are slightly different as the genome sequence has changed between the two releases (Celniker et al. 2002). The ecores created using release 3 were mapped on the collection of gene models defined by the annotations of full-length cDNAs designated as the “Drosophila Gene Collection” (Stapleton et al. 2002; we used a subset of 6,006 transcripts as explained in the Methods section). We only considered ecores located between the start and the end positions of the models. We detected ecores in 87.7% of the genes and in 57.7% of the exons. Six hundred thirty-seven (3.2%) ecores mapped outside the boundaries of annotated exons, and may correspond to alternative exons, nested genes or false positives. In other words, the specificity in this large set was higher than 96%. The mean number of ecores per gene was equal to 3.22 when we considered only ecores overlapping exons, and to 3.33 when considering all ecores within a gene model. Applying these ratios of ecores per gene to the whole genome provides a gene number estimate in Drosophila between 14,036 (46,742/3.33) and 14,516 (46,742/3.22).
Table 1.
Distributions of Ecores in the Sequence of Drosophila in Two Successive Annotations
| Set | Ecores | Genes | Genes detected | Ecores within genes | Exons | Exons detected | Ecores overlapping exons | Ecores overlapping genes not overlapping exons | Ecores/gene |
|---|---|---|---|---|---|---|---|---|---|
| BDGP Release 2 (number) | 47,134 | 13,468 | 11,147 | 42,633 | 54,771 | 31,751 | 41,332 | 1072 | 3.17 |
| BDGP Release 2 (%) | n.d. | n.d. | 82.8 | 90.5 | n.d. | 58.0 | 87.7 | 2.3 | n.d. |
| BDGP Release 3 (number) | 46,742 | 13,666 | 11,167 | 43,705 | 61,085 | 33,996 | 42,679 | 1026 | 3.2 |
| BDGP Release 3 (%) | n.d. | n.d. | 81.7 | 93.5 | n.d. | 55.7 | 91.3 | 2.2 | n.d. |
Genes and exons stand for annotated genes and exons in the corresponding versions
Ecores were also compared to the two last BDGP (Berkeley Drosophila Genome Project) genome annotations (http://www.fruitfly.org/annot/release2.html and http://www.fruitfly.org/annot/release3.html, Misra et al. 2002; Table 1). We observed a significant increase in the percentage of ecores falling inside gene models between the two releases (93.5% versus 90.5%). This provides an independent verification of the improvement of the D. melanogaster annotation between the two versions.
The gene number estimate is based on a ratio of ecores per gene determined using existing annotations and, as a consequence, could reflect a bias in this set. This bias would in particular depend on the level of sequence conservation of genes and on their structure (length and number of exons). However, the collection of 6006 full-length cDNAs from the Drosophila Gene Collection is based on biologic observations, and hence considered as representative. Altogether, these genome comparisons reveal the presence of 4063 ecores outside of annotated exons in the Drosophila genome. Because the mean ecore number in the Drosophila Gene Collection is higher than in other annotated genes, we expect that some gene models are still incomplete or fragmented. We expect that most of these would correspond to additional exons of partially annotated genes. Conversely, it is not expected that these 4063 ecores will contribute to a substantial increase in the total gene number of Drosophila. A verified example of a modification of a predicted gene indicated by Exofish is shown in Figure 1. In this case, a series of additional exons in the release 2 annotation is predicted by Exofish, suggesting that a significant number of exons were missed in this region (Fig. 1A). We reexamined the same region in release 3, and observed that after the new annotation, all ecores are placed in two gene models (Fig.1B). A second example is seen in Figure 2, where the presence of two ecores in a region without annotation in the two insect genomes revealed the existence of a totally new gene, confirmed by a spliced mosquito cDNA.
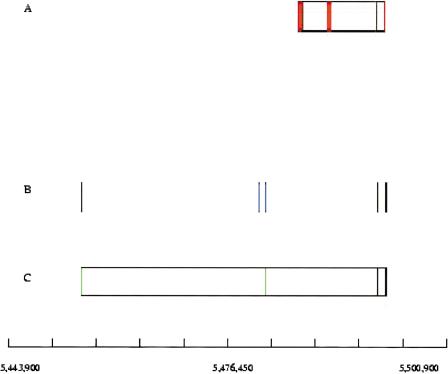
Figure 1.
Exofish analysis on a region on arm 2L of the genome of Drosophila from two different releases of annotations, and around the same ecores. (Top) Results from release 2 of BDGP. (Bottom) Results from release 3 of BDGP. (A, D) BDGP annotations on the 5′-3′ strand. (B, E) BDGP annotations on the 3′-5′ strand. The genes are represented by boxes, with vertical lines separating exons (red) and introns (white). (C, F) Ecores (blue). In release 2 (top), five ecores (numbers 7, 8, 9, 11,12) overlap four gene models, and seven ecores (numbers 1, 2, 3, 4, 5, 6, 10) do not overlap any annotation. In release 3, a large gene model overlaps all the ecores that fall exclusively in exons except ecore number 9. This ecore is part of a gene model on the 5′-3′ strand, which is predicted inside one intron on the 5′-3′ strand.
Figure 2.
Ecores detecting a new gene model. The scale refers to the position on the chromosome arm 2L of the genome of Anopheles. (A) Ensembl gene predictions on the 5′-3′ strand. (B) Ensembl gene predictions on the 3′-5′ strand. The genes are represented by boxes, with vertical lines separating exons (red) and introns (white). (C) Ecores (blue). (D) A confirmatory cDNA sequence is in green, with a potential intron in white. Only one cDNA, matching with two consecutive unanotated ecores, is represented here. This cDNA (corresponding to the assembly of entries BX034944 and BX034945) matches a region unannotated in both Drosophila and Anopheles genomes.
We also ran Exofish against the additional 1042 candidate genes recently proposed for Drosophila (Gopal et al. 2001; http://genomes.rockefeller.edu/dm). We obtained ecores on 18.7% of these new gene models (the list of the matches can be found at www.genoscope.cns/externe/Fly). This low fraction could result from a very low conservation of these genes between Anopheles and Drosophila, possibly representing a subset of rapidly evolving genes, or from a substantial number of false-positive predictions. However, Exofish can serve to validate a number of these potential genes.
The Anopheles Annotation
We also attempted to use Exofish in a reverse mode to identify ecores in Anopheles, assuming that if one ecore in the genome of Drosophila flags a coding sequence, the corresponding ecore in Anopheles should flag a coding sequence. To test the reverse mode, we applied Exofish to a 585-kb region from the Pen1 locus of Anopheles using the whole genome of Drosophila. This region had been independently annotated manually (unpublished results). We detected 100 ecores in this region, with only six of them lying outside of annotated exons, while 83% of the annotated genes are confirmed by at least one ecore. This shows that the expected sensitivity of Exofish should be comparable in this reverse mode. A genome-wide analysis was then performed with the whole A. gambiae assembly.
We found more ecores in the Anopheles assembly (54,069 for release 6.01a) than in the Drosophila genome (ratio = 1.16). The mean size of the ecores is identical for both species (251 nucleotides). Several explanations that are not mutually exclusive may account for this observation. The high number of ecores could be the consequence of (1) an increased coding capacity in the genome of Anopheles, or (2) a larger number of pseudogenes or unmasked tranposable elements in Anopheles, or (3) problems in the sequence assembly. Explanations (1) and (2) were not supported by a previous comparative analysis (Zdobnov et al. 2002). The presence of at least two different haplotypes in the A. gambiae strain sequenced is known to have introduced a number of redundancies in the assembly, essentially as linked artefactual duplications and unanchored duplicated scaffolds (Holt et al. 2002). We analyzed the redundancy in both genomes looking for multiple occurrences of two ecores in one genome created by a single common region in the other genome. A striking result was observed for the alignments occurring once in Drosophila and twice in Anopheles (n = 3476), which were more abundant than the reverse (once in Anopheles and twice in Drosophila, n = 1650, see Methods). We observed significantly more duplicated ecores in the same chromosome in Anopheles (77% of the duplicated cases) than with Drosophila (60%). One exception was noted for chromosome X, where duplicated ecores have their second copy randomly present in the Anopheles genome. This corresponds to the expectation, because chromosome X is the only Anopheles chromosome assembled essentially from a single haplotype (Holt et al. 2002), apparently because of selection in the sequenced strain. An even more striking result is obtained when looking at small, unmapped scaffolds. These sequences represent only 16% of the size of the whole assembly, but contained about 35% of the duplicated ecores. Taken together, these results indicate that an important fraction of the excess ecores resides in regions with potential assembly problems. Further improvements of the A. gambiae genome annotation will be greatly dependent on resolution of the misassembled regions.
We compared the 54,069 ecores from the assembly of Anopheles to release 6.1a of the Celera-Ensembl joint annotations of Anopheles (http://www.ensembl.org/Anopheles_gambiae). We found that 79% of the ecores matched with 79.1% of the gene candidates (Table 2). The fraction of annotated Anopheles genes that is detected by Exofish is thus slightly lower than in Drosophila. Conversely, a large fraction (21%) of Anopheles ecores map outside of annotations. These observations indicate that a substantial fraction of exons were not annotated and that a number of gene models should be revised.
Table 2.
Comparisons Between Ecores on the Assembly of Anopheles and the Successive Ensembl Annotations
| Set | Ecores | Genes | Genes detected | Ecores overlapping genes | Exons | Exons detected | Ecores overlapping exons | Ecores overlapping genes not overlapping exons |
|---|---|---|---|---|---|---|---|---|
| EnsEMBL Release 6.1a (number) | 54,069 | 15,088 | 11,929 | 42,693 | 53,693 | 32,553 | 40,278 | 2,415 |
| EnsEMBL Release 6.1a (%) | n.d. | n.d. | 79.1 | 79.0 | n.d. | 60.6 | 74.5 | 4.5 |
| EnsEMBL Release 10.2.1 (number) | 53,132 | 14,658 | 10,759 | 39,749 | 56,573 | 32,610 | 39,247 | 502 |
| EnsEMBL Release 10.2.1 (%) | n.d. | n.d. | 73.4 | 74.8 | n.d. | 57.6 | 73.9 | 0.9 |
Genes and exons stand for annotated genes and exons in the corresponding versions
A new version of the Anopheles assembly and annotation was recently released (version 10.2.1). This new version addressed some misassembly problems and corrected a number of automatic gene predictions using recent data. Surprisingly, the percentage of ecores outside of annotation increased from 21%–25.6% (Table 2). However, an improvement between the two versions was seen at the level of the redundant ecores. We found that a significant fraction of the duplicated ecores that were present in the release 6.1a have been discarded as haplotype variants. This explained in large part the net disappearance of 937 ecores between the two versions.
Three main types of annotation problems were observed that remained in the two versions. They are exemplified here: absence of annotation in both genomes (Fig. 2); absence of annotation in Anopheles of a known gene in Drosophila (Fig. 3); incorrectly predicted gene lacking some exons and integrating incorrect ones (Fig. 4). In the three examples shown in the figures, the ecores were confirmed by the existence of Anopheles cDNA clones.
Figure 3.
Ecores defining a new gene model on A. gambiae chromosome 2R. The scale refers to the position on the chromosome. (A) Ensembl gene predictions on the 5′-3′ strand. The genes are represented by boxes, with vertical lines separating exons (red) and introns (white). (B) Ecores (blue). (C) Anopheles cDNA clone (green), with potential introns in white. Only one cDNA, matching with three consecutive unanotated ecores is represented here. This cDNA (corresponding to the assembly of entries BX063894 and BX063895) matches all along its sequence with the Drosophila Innexin-7 gene. This gene is not annotated in both releases of the Anopheles annotation.
Figure 4.
Ecores correcting a gene model. The scale refers to the position on the chromosome arm 3L of the genome of Anopheles. (A) Ensembl gene predictions (release 6.1a) on the 5′-3′ strand. The genes are represented by boxes, with vertical lines separating exons (red) and introns (white). (B) Ecores (blue). (C) A cDNA sequence is in green, with potential introns in white. Only one cDNA, matching with unannotated ecores, is represented here. This cDNA (corresponding to the assembly of entries BX062803 and BX062804) matches two of the three orphan ecores. It is homologous throughout to a Drosophila tetraspanin family member. The version 6.1a of the annotation apparently fused the two last exons of the gene with two putative exons, originating from a transposable element. The large sizes of the two first introns may induce such erroneous model constructions. In release 10.2.1, the region is entireley unannotated.
This study shows how whole genome comparisons based on a tool like Exofish can be used as an efficient method to evaluate the quality and to improve existing annotations of insect genomes. In particular, it provides an independent assessment of the improvement of the Drosophila annotation across the successive releases. The fact that 4,063 ecores do not overlap annotated Drosophila exons illustrates the potential of interspecies comparisons, even for extensively studied species like Drosophila. The number of ecores outside annotations in A. gambiae (13,791; Table 2) is higher than for Drosophila, showing that the present automated annotation is probably missing a substantial number of coding sequences. Two successive versions of the annotation gave globally comparable results, reflecting the slow progress in the acquisition of functional and comparative data for annotating this organism. Anopheles/Drosophila ecores can clearly serve to refine and improve the next versions of the Anopheles annotation. The precise locations of ecores in each genome are available for improving both annotations (http://www.genoscope.cns.fr/Exofish/Fly). More generally, this study illustrates the power of whole-genome comparisons, and could be extended to other species combinations with the availability of newly sequenced genomes.
METHODS
Exofish Procedure
To determine the conditions that would generate alignment in coding regions, we first tested a large range of TBLASTX (Altscul et al. 1990) conditions (W,X, scoring matrix) between the ADH region of Drosophila that contains 222 transcripts (Ashburner et al. 1999), and a collection of 16 Mb of shotgun reads from the Anopheles genome. All sequences were masked against known repeats. For each condition, we kept an alignment if all of the alignments with the same length and percent identity were located in a coding region. We selected the conditions that provided the highest sensitivity (match score = 15, mismatch score = -3, W = 4, X = 19). We created a general filter based on the combination of length and percent identity that distinguish alignments falling exclusively in exons from others. For this purpose, we added a collection of sequences of 591 introns of chromosome X of Drosophila (Benos et al. 2001) to the ADH region. We compared this resource to a collection of 310 Mb of shotgun reads from the Anopheles genome. Applying these criteria a series of alignments was selected. We joined overlapping alignments to form ecores. Exofish is a three-step process: compute alignments/filter/create ecores.
Reverse Mode and Ecores Duplicated
Ecores can be built either on the sequence of one species, or on the sequence of the other one among the two genomes being compared (reverse mode). We can link one ecore on one genome to one ecore (eventually more than one) on the other genome if they have common local alignments. To investigate duplications, we selected situations where one ecore on one genome is linked to two ecores (on the other genome) that are both exclusively linked to the same ecore.
Selection of a Drosophila Reference Gene Set
To have a good estimate of sensitivity and specificity of Exofish, we needed a collection of nonredundant and complete genes. We choose the BDGP gene models that correspond to a DGC reference (Stapleton et al. 2002). We eliminate the genes that have at least one intron overlapped by another annotation of the BDGP from this set. Hence, we retained 6006 gene models.
Computations
Anopheles cDNA were mapped on the genomic sequence using Sim4 (Florea et al. 1998).
The series of BLAST comparisons were performed using the Lassap package (Glemet and Codani 1997). All the computations were performed on a cluster of 40 CPU α (EV6.8/1GHz) organized in eight nodes (7 ES45 + 1 GS160-12) using the Cluster File System.
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.922503.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Misra, S., Roote, J., Lewis, S.E., Blazej, R., Davis, T., Doyle, C., Galle, R., George, R., Harris, N., et al. 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics 153: 179-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos, P.V., Gatt, M.K., Murphy, L., Harris, D., Barrell, B., Ferraz, C., Vidal, S., Brun, C., Demaille, J., and Cadieu, E. 2001. From first base: The sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 11: 710-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., and Frise, E. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: 7901-7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glemet, E. and Codani, J.J. 1997. LASSAP, a LArge Scale Sequence compArison Package. Comput. Appl. Biosci. 13: 137-143. [DOI] [PubMed] [Google Scholar]
- Gopal, S., Schroeder, M., Pieper, U., Sczyrba, A., Aytekin-Kurban, G., Bekiranov, S., Fajardo, J.E., Eswar, N., Sanchez, R., Sali, A., et al. 2001. Homology-based annotation yields 1,042 new candidate genes in the Drosophila melanogaster genome. Nat. Genet. 27: 337-340. [DOI] [PubMed] [Google Scholar]
- Holt, R.A., Subramanian, G.M., Halpern, A., Sutton, G.G., Charlab, R., Nusskern, D.R., Wincker, P., Clark, A.G., Ribeiro, J.M., Wides, R., et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: 8301-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural, R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1161-1171. [DOI] [PubMed] [Google Scholar]
- Roest Crollius, H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 25: 235-238. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. 2002. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E.M., Von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.fruitfly.org/DGC; BDGP; Drosophila gene collection.
- http://www.fruitfly.org/annot/release2.html; BDGP; Drosophila genome annotation release 2.
- http://www.ensembl.org/Anopheles_gambiae; ENSEMBL mosquito genome server.
- http://www.genoscope.cns.fr/Exofish/Fly; Genoscope Anopheles/Drosophila Exofish database.
- http://genomes.rockfeller.edu/dm; A collection of additional candidate genes in Drosophila.