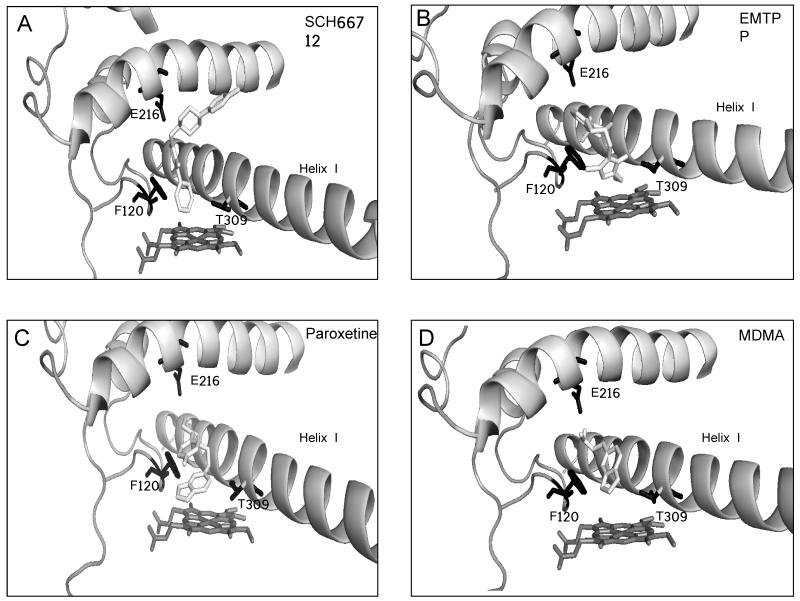
Figure 4.
Molecular modeling of inactivators bound to CYP2D6. AutoDock was used to model binding in the active site of each inactivator in the conformation believed to lead to inactivation of CYP2D6 as described in the Materials and Methods. Docking simulation with (A) SCH66712, (B) EMTPP, (C) paroxetine, and (D) MDMA. Active site amino acids Phe120, Glu216, and Thr309 are shown in black.