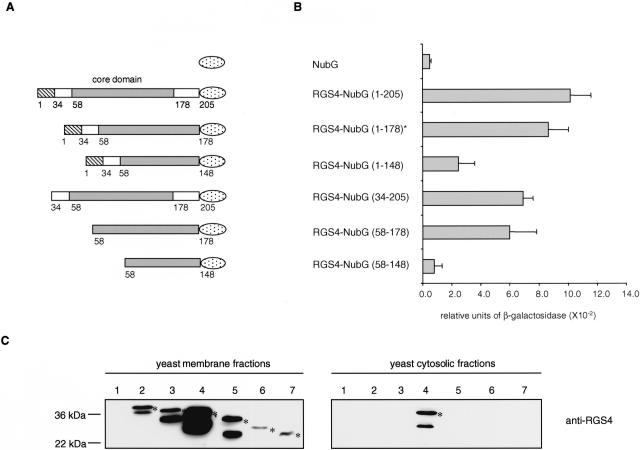
Figure 5.
Mapping of the ErbB3 interaction domain within RGS4. (A) RGS4-NubG deletion constructs. RGS4 contains an N-terminal amphipathic region important for membrane localization (amino acid 1–33, striped box) followed by the RGS box (amino acid 58–178, grey box) and the C-terminal tail (amino acid 179–205). The NubG domain of ubiquitin is shown as a dotted ellipse. (B) Quantitative β-galactosidase assay showing the binding of different RGS4-NubG deletion mutants to the ErbB3-Cub-TF bait. Values given are the mean of four transformants, each assayed at least twice. The asterisk indicates the original RGS4 clone isolated in the split-ubiquitin membrane yeast two-hybrid screen. (C) Expression and localization of the RGS4–2HA-NubG deletion mutants. The RGS4–2HA-NubG deletion mutants were generated by PCR followed by in vivo recombination into the prey plasmid pADH-PREY-2HA-NubG previously digested with NdeI. Western blot analyses were performed using membrane (left panels) and cytosolic (right panels) fractions of the yeast L40 reporter strain expressing ErbB3-Cub-TF and the prey NubG (lane 1), RGS4-NubG (1–178; lane 2), RGS4-NubG (1–205; lane 3), RGS4-NubG (34–205; lane 4), RGS4-NubG (1–148; lane 5), RGS4-NubG (58–178; lane 6), and RGS4-NubG (58–148; lane 7). The blots were incubated with anti-RGS4 antibody. The positions of molecular markers are indicated. The bands that are not marked with an asterisk are considered as degradation products of RGS4. The RGS4-NubG (1–178) protein found in the split-ubiquitin membrane yeast two-hybrid screen (lane 2) migrates more slowly than the RGS4-NubG (1–205; lane 3) because it contains an additional 45 amino acids upstream of the initial RGS4 start codon.